9 Regulation of Gene Expression
Walter Suza; Donald Lee; Philip Becraft; and Marjorie Hanneman
- Understand the concept of gene expression.
- Understand transcriptional regulation of gene expression.
- Understand epigenetic gene regulation.
- Understand post-transcriptional regulation of gene expression (RNA level).
- Understand post-translational protein modification and regulation.
Introduction
Every cell in a plant contains the same genetic information, the same set of genes. Yet Therefore different sets of genes are required for the various functions of different cells or tissues, as well as for plant responses to environmental stimuli or stresses. This is achieved by regulating the activity of genes according to the physiological demands of a particular cell type, developmental stage, or environmental condition. This regulation of activity is known as gene expression.
The term expression can be used in different ways that are sometimes confusing. Typically, if a gene product is produced, the gene is considered “expressed”. However, it sometimes occurs that a transcript might be produced but not a protein, or that a protein is produced but it is in an inactive state. In such cases, although a gene product is produced, the biological activity encoded by that gene is not present. For the purposes of this section, the key point is how the biological activity encoded by a gene is regulated.
The expression of genes in specific plant cells, tissues, and organs and the timing of this expression require a precise level of regulation. Expression, or genetic function, can potentially be regulated at any of the steps from transcription, RNA processing, translation, through post-translational protein modification, as discussed in lesson 1. Regulation can be qualitative (i.e. gene expression is either “on” or “off”) or quantitative (i.e. expression levels can be modulated “up” or “down”). Fluctuations in the intensities of external stimuli coupled to changes that occur at the genomic level result in different developmental outcomes or physiological states. Regulation of gene expression at the level of transcription can be brought about through chromatin and histone modifications. Also, a gene sequence can be differentially spliced to produce mRNA products of variable lengths leading to new protein products with novel functions. Some genes do not encode proteins but short forms of RNAs with regulatory functions such as induction of flowering. Finally, proteins products can be subjected to modifications such as phosphorylation or dephosphorylation to alter their functions, or can be completely degraded to turn off a gene.
Transcriptional gene regulation
Since transcription is the first step in gene expression, it makes sense in terms of cellular economy, to regulate expression at this point, and in fact this is one of the most important regulatory points. We already described the involvement of RNA polymerase in the transcription process, but in fact there are other protein factors that are required. Proteins involved in transcriptional regulation are known as transcription factors. It is the interaction of these transcription factors with specific DNA sequences that regulate the process of gene transcription.
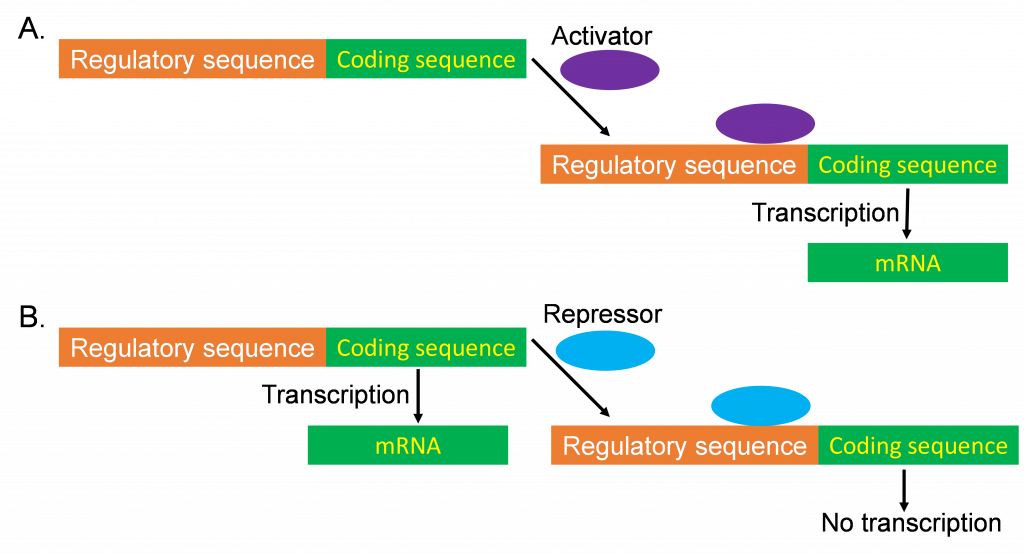
A. The concept of differential (regulated) gene expression
As just described, not every gene is expressed all the time. When a gene displays different levels of expression in different circumstances, this is known as differential expression. Circumstances that might apply include, but are not limited to, different plant tissues (root vs. leaf), different developmental stages (germination vs. reproductive development), or in response to different environmental stimuli (cold stress or pathogen attack).
The term differential expression can also be used to compare the expression of different genes. If two genes show different expression patterns (among plant tissues or in response to environmental stimuli), they are considered differentially expressed, whereas genes that showed very similar patterns of expression would be considered co-expressed.
B. Promoters
As mentioned, transcription is regulated through the interactions of proteins, transcription factors, with specific DNA sequences. Most regulatory DNA sequences governing gene transcription are located on the 5′ border of the transcribed region. This region is called the gene promoter.
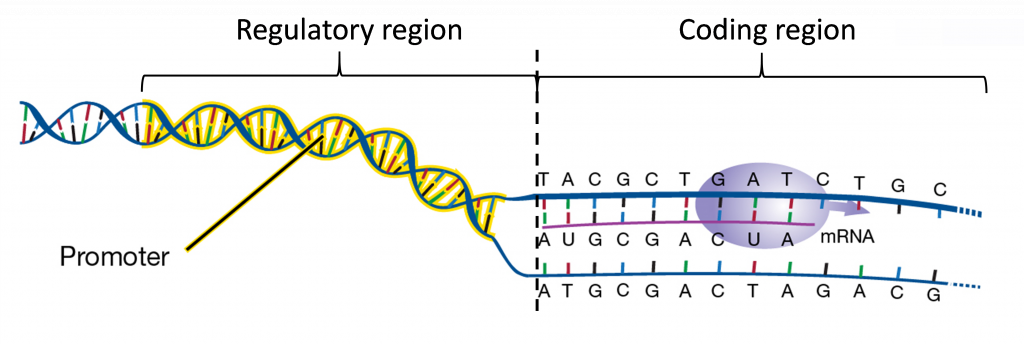
Promoters contain a core, which is required for the binding of the “basal transcriptional machinery”, including RNA polymerase. The “TATA” box, with a consensus sequence TATAA, is located within the core promoter, usually 25-30 nucleotides upstream of the transcription initiation site. Promoters also contain regulatory sequences that determine when, where, and to what level genes are transcribed. Promoters can vary in length from a hundred to a few thousand nucleotides.
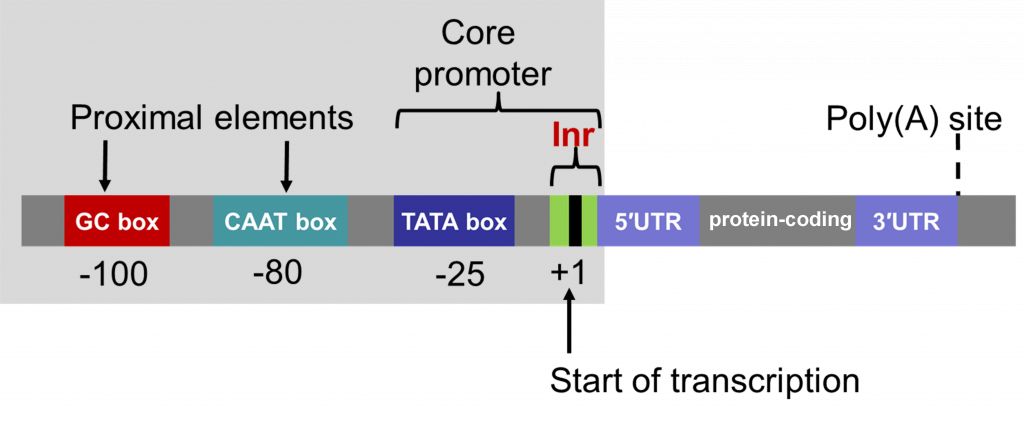
The full promoter sequences of different genes that are expressed in a similar manner may be different. However, such promoters often contain short sequence “motifs” that are similar, referred to as cis elements. Early work (Benfy and Chua, 1990) to understand the function of different promoter elements in regulating gene expression in plant cells and tissues revealed that various combinations of cis elements are able to be interpreted by the cell and control gene expression. Sometimes cis elements promote gene transcription and sometimes they function to restrict transcription of genes in particular cells and tissues.
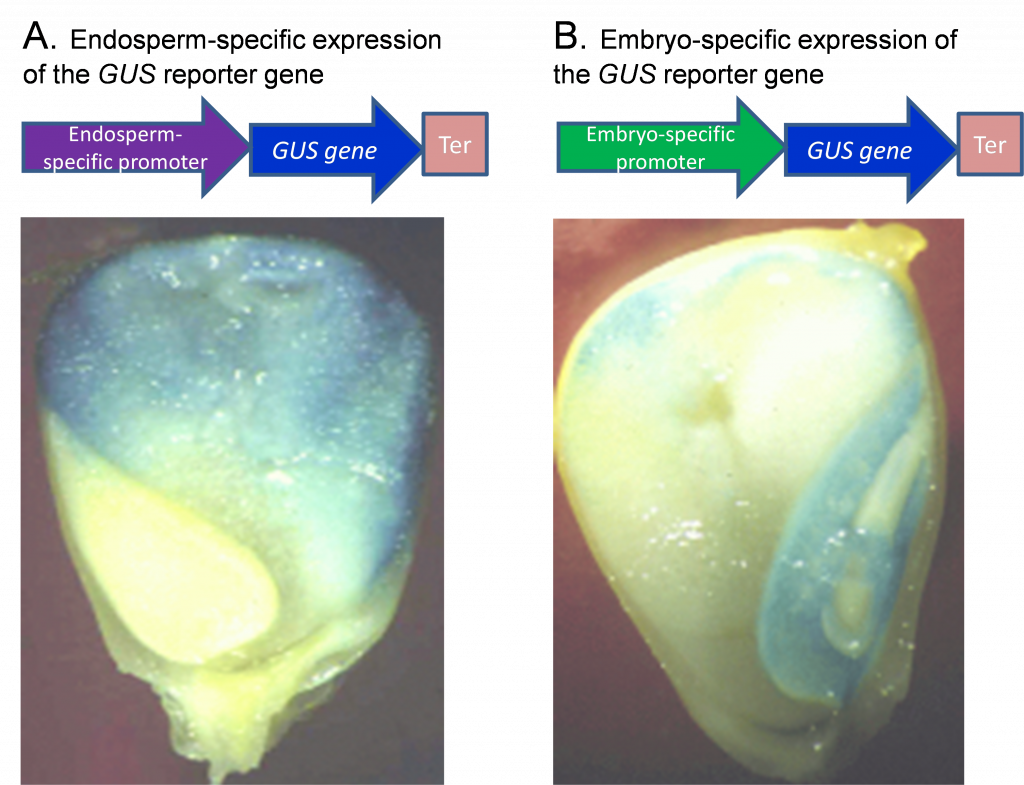
Promoter analysis is facilitated through the use of reporter genes. The reporter gene produces effects that are easily identifiable and quantifiable, which can be used to determine the function of a regulatory region of another gene (promoter, promoter elements, or enhancers) in cells, tissues, or organs. Such analyses are critical to crop biotechnology where targeted expression of genes in particular tissues is often desirable. To test whether a promoter is effective in conferring the expression of a gene to a particular tissue, scientists fuse the putative promoter to a reporter gene and introduce the promoter-gene fusion into plants. An example of a reporter is the GUS gene, which encodes a GUS enzyme (beta-Glucuronidase) that, when expressed, produces a blue color upon addition of a substrate. Figure 1 shows an example of GUS expression in maize seed using two promoters that act either in the endosperm or the embryo.
C. Enhancers
Enhancers are DNA sequences that increase the rate of transcription of a gene when they are present, although alone they cannot cause transcription to occur. Enhancers are usually position and orientation independent. Although they are normally located upstream of the promoter, they can also be located on the 3’ region of the gene or even with the coding region. Enhancers can increase the transcription when added to genes that they are not normally associated with. This is a useful property for biotechnology, allowing promoters to be manipulated for increased levels of transcriptional regulation. Some enhancers function at all times in all cells and tissues and they are referred to as constitutive. Other enhancers function in specific tissues at specific developmental stages. Some are active only in response to environmental signals. The AACCA enhancer on the promoter of soybean β-conglycinin gene encoding a seed storage protein functions specifically in seeds. Enhancers are thought to interact with specific nuclear proteins involved in transcription. For example, enhancers might facilitate the binding of transcription factors and direct these factors along the DNA strand to the direction of the promoter. Alternatively, enhancers may facilitate changes in DNA structure such as modification of the chromatin structure.
The counterpart of an enhancer is a silencer. Silencers have all the properties just described for enhancers, except they function to dampen, or decrease, the levels of transcription controlled by a promoter.
D. Transcription factors
RNA polymerase binds the promoter at the TATA box and in cooperation with other proteins drives gene transcription. The proteins that interact with RNA polymerase to facilitate its binding to the promoter and to regulate its activity are known as transcription factors. Some transcription factors, known as “basal transcription factors” are fundamental to RNA polymerase binding and function, and are expressed in all living cells.
Other transcription factors bind to cis regulatory DNA sequences of promoters, enhancers or silencers. These proteins interact in complex ways with the basal transcription machinery, to regulate the activity of RNA polymerase, and therefore gene transcription. Thus, transcription factors regulate when or where individual genes are expressed, and to what level.
Transcription factors can function as either positive or negative regulators. That is, they can either function to induce (increase) gene transcription or to repress it. The consequence of many transcription factors depends on their interactions with other proteins. Two factors together may be required for gene activity, and exclusion of one of the factors in space and time offers a mechanism for differential gene expression. Some transcription factors might function as a positive regulator in one context but as a negative regulator in another context, depending on what other cis elements and/or transcription factors might be present.
Epigenetic regulation
A. Chromatin structure and histone modification
At the molecular level chromatin is a product of an ordered and tight packaging of the double stranded DNA around nucleosomes (a core of proteins named histones), and an association with additional proteins.
Transcriptional regulation often involves modification of chromatin structure mediated by post-translational regulation (changes in acetylation and methylation) of histones. Histone acetylation involves the addition of acetyl groups and when histones are heavily acetylated the DNA is less tightly associated with them. This often correlates with increased transcriptional activity of specific genes. The idea is that when DNA is loosely associated with histones, it is more accessible to transcription factors that require interaction with the DNA to initiate transcription. Consequently, histone deacetylation (removal of acetyl groups) by histone deacetylase enzymes (for example, FLD, p462) stabilizes nucleosomes and represses transcription. On the other hand, histone acetylation by histone ecetyltransferase destabilizes nucleosomes and promotes transcription.
As described in the text, another form of histone modification is the addition of methyl groups to histone proteins, which similarly regulates chromatin condensation or decondensation.
B. DNA methylation
The nucleotide bases of DNA can be modified by the attachment of methyl groups at various locations. The addition of these methyl groups happens after the DNA has been synthesized and is controlled by enzymes that add methyl moieties to specific regions of DNA. The most common modified nucleotide base is C5 methylcytosine (m5C) due the activity of the enzyme DNA (cytosine-5) methyltransferase (MET). MET recognizes the unmethylated newly-replicated DNA strand and incorporates a methyl group if the template strand was methylated (hemimethylated).
Methylation status is correlated with gene expression (Figure 5) as demonstrated by the low level of methylation in regions of the genome undergoing active transcription.
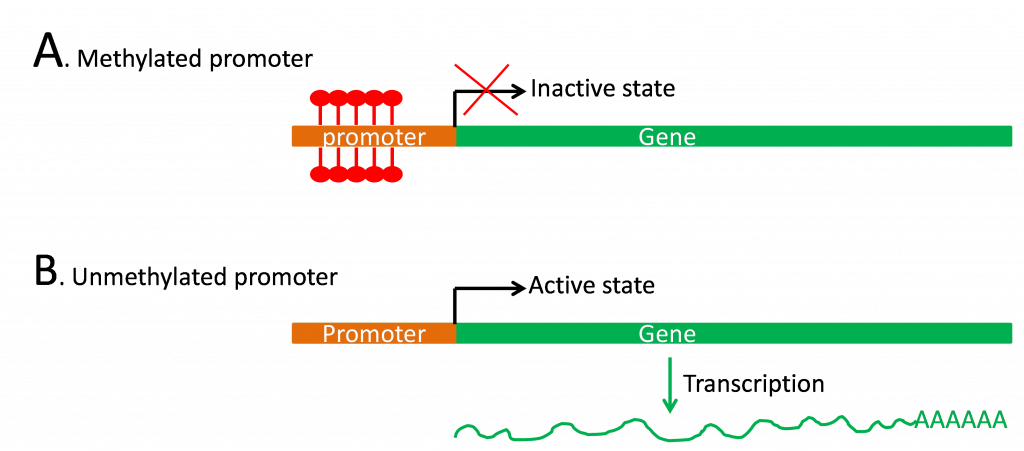
Studies have shown that altering a plant’s overall DNA methylation status can affect growth and development. For example, cold treatment (vernalization) induces flowering in biannuals such as Arabidopsis and winter wheat, and lowers the level of methylation of particular genes. Also, treating plants with the drug 5 azacytidine prevents methylation at the 5 position of cytosine and stimulates flowering. Recent studies also suggest a relationship between epigenetic gene regulation and heterosis, with hybrids showing higher global levels of transcription, higher histone acetylation levels and lower DNA methylation levels (reviewed in He et al., 2011).
RNA-level regulation
Regulation of gene expression occurs at many levels, including post-transcriptionally. From the standpoint of the expression level for a given gene, a critical factor is the level of fully processed, mature mRNA. When we consider the level of the mature form a particular transcript at any given time reflects the steady state balance between synthesis, processing and degradation. The Post-transcriptional regulation of RNA occurs through several mechanisms.
RNA stability and degradation
The control of mRNA degradation rate, or turnover, is an important regulatory mechanism in gene expression. As mentioned, the level of a particular transcript at any given time reflects the steady state balance between synthesis and degradation. Thus, even though a gene may be transcribed at a high rate, a high rate of RNA turnover could effectively shut off the gene. The stability of mRNA can be regulated globally, to affect all or most transcripts, or it can be very specific to a particular mRNA. Regulated mRNA turnover may be particularly important in plants, since plants cannot move to avoid harsh environmental conditions and stresses are known to induce specific changes in RNA stability. Other factors such as light and plant hormones are also known to regulate mRNA stability.
The degradation of mRNA is catalyzed by enzymes called ribonucleases. Ribonucleases include exonucleases, which degrade only from one end of the transcript, and endonucleases, which can attack the mRNA molecule internally.
Therefore, controlling ribonuclease access to the mRNA substrate is an important regulatory mechanism in gene expression. This control is achieved through various mechanisms including, controlling the amount of nucleases present, regulating the activity of the nuclease, sequestering the nuclease to particular cellular locations to restrict its access to RNA, and to control the stability of the mRNA by decreasing accessibility to nuclease.
Four regions of an mRNA are important for its overall stability. These include, the 5′ untranslated region and the cap, coding region, 3’ untranslated region, and the poly(A) tail.
The cap at the 5′ end protects the mRNA from exonucleases that degrade RNA from the 5′ to the 3′ direction.
The 3′ untranslated region may contain short sequences that influence mRNA stability. For example, the repetition of the sequence AUUUA in the 3′ untranslated region of many animal and plant genes is associated with mRNAs with short half-lives (the time it takes for half the RNA to be degraded, after transcription is stopped).
The poly(A) tail increases mRNA stability but is not sufficient alone. It only provides stability when bound by proteins such as poly(A) binding protein (PABP). Test-tube experiments have shown that, removing PABP from a stable mRNA destabilizes it, while adding back purified PABP restores stability.
Translational regulation
As described, for most genes it is the protein product that performs the biological function. As such, regulating the amount of protein production effectively regulates gene expression. There are several known mechanisms by which the process of translation is known to be regulated. A detailed consideration of these mechanisms is beyond the scope of this course, but in general it is important to bear in mind their importance. For example, under certain stress conditions, “normal” translation is halted and only mRNAs related to stress tolerance are selectively permitted to be translated into proteins. This selective translation is important to allow plants to quickly respond to stress and to conserve energy under stress conditions.
Translation of mRNAs can play an important role in determining their overall stability. Mutations that add premature stop codons often lead to rapid degradation of the mRNA. Ribonucleases (figure 9) or other factors involved in degradation may recognize the number or spacing of ribosomes on an mRNA, and degrade those that are not produced properly. This may help prevent synthesis of proteins with incorrect functions which will negatively impact cellular processes. Errors during transcription could add or omit nucleotides which would alter the proper codon sequence creating mutant proteins.
Protein-level regulation
After translation, proteins are subject to a variety of modifications that can regulate their activity. There are many different ways by which proteins can potentially be modified and thereby regulated, and only a few of the basic mechanisms will be considered here.
Common types of post-translational modifications
The first mechanism is covalent modification by the addition of various chemical groups. A wide variety of groups can be involved, including small organic groups (methylation, acetylation) lipids (myristoylation, farnesylation, palmitylation), carbohydrates (glycosylation, glucosylation), small proteins (ubiquitination, sumoylation) and inorganic molecules (phosphorylation, sulfation). Such covalent modifications are generally accomplished by the activity of enzymes specialized to perform these modifications. Many of these modifications are also reversible; that is these groups can be added to a protein and subsequently removed. Phosphorylation is a particularly noteworthy reversible modification that is common in the regulation of many proteins. Enzymes that add phosphate groups to other proteins are called protein kinases, and those that remove phosphates are called phosphatases. As such, protein kinases and phosphatases are central to many cellular regulatory systems.
A second common mechanism of protein modification is through proteolytic cleavage. Proteolysis occurs as part of the general turnover of cellular proteins, which is required to eliminate damaged proteins and recycle amino acids. Proteolysis is important for the processing of certain proteins, for example in the removal of the signal peptide of proteins targeted to specific cellular compartments. It can also occur in a highly specific manner whereby particular proteins are targeted for degradation or for cleavage at a specific site within the protein. Proteolytic modifications are non-reversible.
Proteins can undergo modification through complex formation. Such complexes can occur among proteins or between a protein and a cofactor.
Proteins can also be modified according to conditions of the cellular environment. The redox state can result in oxidation or reduction of proteins, particularly of sulfhydryl side groups. Cellular pH can affect the charge of ionizable side groups.
Common types of protein regulation
All of the above mentioned types of protein modification can alter protein conformations and thus have regulatory consequences on protein function or activity. Protein regulation is highly complex and there are a myriad of different ways by which this occurs. Again we will just briefly consider a few of the more common mechanisms.
One major way protein modification can regulate protein function is by altering their activity. This can be true for many types of proteins including, but not limited to, enzymes, transcription factors, signaling proteins, and structural proteins.
Cells are compartmentalized into several membrane bound organelles, including the nucleus, chloroplasts, mitochondria, peroxisomes, endoplasmic reticulum (ER), golgi, and vacuoles. Each of these compartments performs unique metabolic functions that require a set of proteins. Compartmentation is regulated for some proteins. For example, upon exposure to light the phytochrome protein moves into the nucleus where it affects the expression of light-regulated genes.
Proteolytic processing is involved in several important regulatory processes. Many proteins are synthesized in an inactive form that requires proteolytic cleavage for activation. The full-length translation product before processing is often called the precursor, or a preprotein. As mentioned, cells contain membrane-bound compartments. Since membranes are impermeable to most proteins, an active mechanism is necessary to move a protein across a membrane. For proper delivery to their organellar destinations, specific amino acid sequences, called target signals must be present in a protein (to serve as an “address”). For example, entry into the chloroplast is achieved by the presence of a target signal called the transit peptide. This is at the amino terminal end of the protein and is proteolytically cleaved during import.
Another example of proteolytic processing is seen in a plant defense response in members of the solanaceae (for example, tomato and potato). An 18-amino acid peptide hormone called systemin is secreted by plant cells that are damaged by insects or mechanical wounding. Systemin production by wounded cells is required to induce the synthesis of proteins involved in defense. Systemin induces defense responses in wounded cells, and throughout the plant. Analogous to animal peptide hormones (e.g., insulin), systemin is initially synthesized as a much larger (200 amino acids) precursor called pro-systemin. Pro-system is inactive; however, upon wounding it undergoes proteolytic cleavage to produce activated systemin.
Targeted protein degradation
The amount of protein present in a cell or tissue is determined by both its rate of synthesis and its rate of degradation. Therefore, protein degradation is an important mechanism by which the plant can regulate biological activity (i.e., a genetic function). For example, one way to shut down a metabolic pathway is by degrading one of the key enzymes controlling the rate of the entire pathway. Therefore, protein degradation is an essential component of gene regulation to meet cellular demands for growth, development, and defense.
Protein degradation must be carefully controlled to fine tune gene expression to allow plants to adapt to new environmental conditions. Often, cells will adopt several complex mechanisms for proteolytic degradation of proteins. Enzymes that cleave or degrade proteins are referred to as proteases. For example, plant vacuoles are rich in proteases that play a similar function in protein degradation as that of lysosomes in animal cells. Protease activity must be tightly regulated to prevent accidental degradation of essential proteins. Sequestering proteases in particular organelles like the vacuole separates them from other organelles and is one of to control their activity.
An important mechanism by which specific proteins are targeted for degradation is through ubiquitin-mediated proteasomal degradation. The proteasome is a large complex of multiple protein subunits that have a protease activity to degrade proteins. Proteins get marked for proteasomal degradation with a small protein called ubiquitin. Ubiquitin is covalently attached to specific proteins in response to environmental or developmental signals. This allows plants to quickly adapt to changing conditions by eliminating proteins whose functions are not advantageous under the new conditions. For example, the photoreceptor phytochrome mentioned above becomes targeted for degradation when light is no longer available. This allows plants to change their physiological functions going from daylight to night conditions. Protein degradation by the proteasome system is also an important regulatory mechanism for plant hormone signaling, for example the signaling of the defense hormone jasmonic acid, and growth hormone gibberellic acid.
Proteins have lifetimes that range from a few minutes to weeks or more. Cells continuously make proteins from and break them down to amino acids. One of the functions of protein degradation is to eliminate aberrant or damaged proteins which could harm the cell. The second function is to facilitate the recycling of amino acids. For example, most of the amino acids required for growth of the seedling are derived from the degradation of seed storage proteins. Conversely, in annual crop plants, many of the amino acids in seed storage proteins are derived from proteins degraded in leaves and other plant parts during senescence.
The expression of genes in specific plant cells, tissues, and organs and the timing of this expression require a precise level of regulation. A single promoter may not be sufficient to regulate the expression of such gene(s) in space and time. Therefore, coding regions with the same function may have different promoters, and such genes are referred to as differentially regulated. Most regulatory sequences governing gene expression are located on the 5’ border of the coding region. Transcription is often initiated between 20 and 60 nucleotides upstream of the ATG start site. Enhancers are usually position and orientation independent. Although they are normally located upstream of the promoter, they can also be located on the 3’ region of the gene or even with the coding region. Enhancers can increase the transcription when added to genes that they are not associated with. RNA polymerase binds the promoter at the TATA box and in cooperation with other proteins drives gene transcription. The proteins that interact with RNA polymerase bind to regulatory sequences upstream and downstream of the transcription site.
Transcription factors can play a regulatory role by determining where individual genes are expressed. Transcriptional regulation often involves modification of chromatin by changes in acetylation and methylation of histone. The nucleotide bases of DNA can be modified by the attachment of methyl groups at various locations. Methylation status is correlated with gene expression. Alternative splicing describes an alternative mechanism of pre-mRNA processing to generate mRNAs that have different combinations of exons. The control of mRNA degradation rate, or turnover, is an important regulatory mechanism in gene expression. RNA interference (RNAi) is a post-transcriptional process involving the degradation of mRNA initiated by the formation of double-stranded RNA (dsRNA) of the target mRNA. Translation of mRNAs can play an important role in determining their overall stability. Mutations that add premature stop codons often lead to rapid degradation of the mRNA. Protein degradation is an essential component of gene regulation to meet cellular demands for growth, development, and defense. The plant can alter the activity of a metabolic pathway, by degrading one of the key enzymes controlling the rate of the entire pathway.
1a. Which of the following could alter gene regulation
- Deleting a promoter
- Altering fertilizer application rate
- Raising the temperature of the greenhouse
- Herbicide application
- Reduce irrigation frequency
1b. The detection of a gene product, for example, RNA or protein at any one moment is a reflection of the “steady state” of the product. Describe the term “steady state” in this context.
2. You are studying expression of a gene responsible for resistance to a pathogen. You cloned the gene and made antibody to detect the protein it encodes by a procedure called Western blot analysis, and the mRNA by a procedure called reverse transcription PCR. Explain the type of regulation from the scenarios in the table.
|
Scenarios |
Not treated with pathogen |
Treated with pathogen | ||
|
|
mRNA |
Protein |
mRNA |
Protein |
|
1 |
Absent |
Absent |
Present |
Present |
|
2 |
Present |
Absent |
Present |
Present |
|
3 |
Present |
Present |
Absent |
Absent |
|
4 |
Present |
Present |
Present |
Absent |
|
5 |
Present |
Present |
Present |
Present |
3. You are studying the expression of a gene controlling height in your crop species. You have cloned the gene and are interested in determining its expression in different parts of the plant. You use the RT-PCR procedure as in the previous problem. After the PCR part, you load products on a gel and observe the following pattern. Explain your results in the context of gene regulation.
References
Benfey, P. N., N. Chua. 1990. The cauliflower mosaic virus 35S promoter: Combinatorial regulation of transcription in plants. Science 16:959-966.
Chung, H. S., G. A. Howe. 2009. A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 21:131-145.
He, G., Elling, A.A., and Deng, X.W. (2011). The Epigenome and Plant Development. Annual Review of Plant Biology 62, 411-435.
ISHIHAMA, N., R. YAMADA, M. YOSHIOKA, S. KATOU and H. YOSHIOKA, 2010 Phosphorylation of the Nicotiana benthamiana WRKY8 Transcription Factor by MAPK Functions in the Defense Response. Plant Cell 23: 1153-1170.
Nakaminami, K., Matsui, A., Shinozaki, K., and Seki, M. (2011). RNA regulation in plant abiotic stress responses. Biochimica et Biophysica Acta (BBA) – Gene Regulatory Mechanisms.
Sheen, J., 1991. Molecular mechanisms underlying the differential expression of maize pyruvate, orthophosphate dikinase genes. Plant Cell 3:225-245.

