Chapter 14: Groundnut Breeding
Teshale Mamo; Arti Singh; Asheesh Singh; Anthony A. Mahama; and Walter Suza
Groundnut or peanut (Arachis hypogaea L. Millsp) is a self-pollinated species belonging to the Fabaceae family. It is considered as the most important food legume crop in continental Africa because of its multiple purpose uses in food, feed, paints, lubricants and insecticides as the seeds are comprised of 35–56% oil, 25–30% protein, 9.5–19.0% carbohydrates, several minerals like P, Ca, Mg and K, as well as vitamins E, K and B (Gulluoglu et al. 2016).The crop has various industrial uses including products such as food, feed, paints, lubricants and insecticides. Because of its natural ability to fix atmospheric nitrogen in the soil, it is considered an ideal crop in crop rotation systems and in intercropping under small scale subsistence farming systems. Plagued by many biotic and abiotic growth limiting factors, production has been lower than the potential. Breeding to alleviate or overcome these limiting factors is therefore key new cultivars to ensure increased productivity to contribute toward assured food security.
- Become familiar with the basic biology and importance of groundnut
- Provide examples of breeding schemes used to develop cultivars of groundnut
Crop Attributes
Crop Biology

Groundnut or peanut (Arachis hypogaea L. Millsp) is a self-pollinated species belonging to the Fabaceae family. Groundnut is a disomic allotetraploid (2n = 4x = 40). The two sets of chromosomes of A. hypogaea are highly diplodized, meaning there is little recombination between the A and B genomes except when the infrequent quadrivalent is formed. Groundnut is found in the Arachis section along with A. monticola, also a tetraploid, and ~25 diploid species (Dwivedi et al., 2007). Arachis is subdivided into nine taxonomical sections: Arachis,Erectoides, Rhizomatosae, Extranervosae, Heteranthae, Trierectoides, Triseminatae, Caulorrhizae, and Procumbentes with groundnut classified in the Arachis section (Dwivedi et al., 2007).
Crop Geography
The genus Arachis originated in South America and is comprised 68 species (Dwivedi et al., 2007; Krapovickas and Gregory, 1994). The species of Arachis are easily delineated from other closely related genera because they flower above ground but set seed below ground (Holbrook and Stalker, 2003).
There are six centers of diversity for groundnut in South America including geographic regions in Paraguay-Paraná, the upper Amazon, the west coast of Peru, Brazil, and the southwest Amazon region in Bolivia. A secondary center of diversity also exists in Africa (Holbrook and Stalker, 2003; Wynne and Coffelt 1982). Arachis hypogaea is believed to have originated in the South American region encompassing southern Bolivia to northern Argentina (Holbrook and Stalker, 2003). Arachis hypogaea is thought to have arisen ~4000 years ago from a single hybridization event between two diploid Arachis species (i.e., A genome from A. duranensis, B genome from A. ipaensis) followed by a spontaneous chromosome doubling of the sterile hybrid to form a fertile allotetraploid (i.e., AABB) (Kochert et al., 1996; Young et al. 1996). Though the fertility was restored in the resulting allotetraploid it was reproductively isolated from its progenitor species creating a strong genetic bottleneck, which is partially responsible for the low allelic diversity present in modern cultivated peanut (Dwivedi et al., 2007; Kochert et al., 1996; Stalker et al., 2013; Young et al. 1996).
Crop Inflorescence
Arachis hypogaea or cultivated groundnut has simple or compound (i.e., 1-5 flowers) inflorescences that occur in the leaf axils on both primary and secondary branches. Typically a single flower per inflorescence opens per day (Holbrook and Stalker, 2003). The flower includes a standard, wing, and keel petals. The standard is deep orange to light yellow in color, and in rare cases may be white. The calyx consists of five sepals attached to the elongated hypanthium. Flowers possess an elongated tubular hypanthium or calyx tube so they look like they are borne on stalks but are instead classified as sessile (Holbrook and Stalker, 2003).
Arachis hypogaea is divided into two subspecies (e.g., hypogaea and fastigiata) and six botanical varieties (e.g., hypogaea, hirsuta, fastigiata, peruviana , aequatoriana, and vulgaris) (Table 1) (Holbrook and Stalker, 2003).
Subspecific and Varietal Classification
| Botanical variety | Market type | Location | Traits |
|---|---|---|---|
| hypogeae | Virginia/Runner | Bolivia, Amazon | No flowers on the central stem, alternating pairs of floral and vegetative axes on lateral branches, short branches, spreading growth habit, relatively few trichomes
Virginia type: large seeds, less hairy, consumed in the shell or roasted, longer growing cycle Runner type: small seeds, less hairy, used to make groundnut/peanut butter |
| hirsuta | Peruvian runner | Peru | more hairy |
| fastigiata | Valencia | Brazil, Paraguay, Peru, Uruguay | Flowers in the mainstem, sequential/disorganized pairs of floral and vegetative axes on the branches, more erect to procumbent, shorter life cycle, pods with 2+ seeds, smooth pericarp, consumed in the shell, boiled, or canned, primary type in Uganda |
| peruviana | n/a | Peru, Bolivia | Less hairy, deep pod reticulation |
| aequatoriana | n/a | Ecuador | Very hairy, deep pod reticulation, purple stems, more branched, upright |
| vulgaris | Spanish | Brazil, Paraguay, Uruguay | More branched, upright, shorter life cycle, flowers on the central stem, floral and vegetative axes arranged in a disorganized fashion, 2 seeds per pod |
Importance
Groundnut is a legume crop adapted to a hot, semi-arid climate, survives and yields under rainfed conditions, fixes nitrogen, and requires few inputs, making it a crop that is suitable for small shareholder farms. It can be grown in a low input environment and provides a source of fat and protein, and provides many phytonutrients including vitamin E and antioxidants.
Usage
Groundnut is a multiuse crop that is harvested as a source of edible oil, vegetable protein, and forage (haulms) for horse and cattle (Dwivedi et al., 2003). Groundnut is a rich source of oil, protein, minerals (Ca, Mg, P, and K), and vitamins (E, K, and B1) (Savage and Keenan 1994). Groundnut contains 40-60% oil, 20-30% protein and, and 1020% carbohydrate (Dwivedi et al., 2003; Pandey et al. 2012; Savage and Keenan 1994)
In the United States, approximately 70 percent of the groundnuts are runners (small-seeded types of var. hypogaea), 20 percent are virginias (large-seeded types of var. hypogaea), 10 percent are spanish (var. vulgaris), and less than 1 percent are valencia (var. fastigiata) market types (Holbrook and Stalker, 2003; Knauft and Gorbet, 1989. Most of the groundnut produced is for human consumption, but low quality lots (blemished seeds, aflatoxin contamination) are crushed for oil.
The quality of groundnut oil is determined by the ratio of oleic (O) to linoleic (L) fatty acids. A higher ratio results in better storage quality of the oil that is less prone to oxidation and the development of undesirable flavors, and has a longer shelf life. For groundnut varieties harvested for oil extraction (i.e., high oil, high O/L ratio) seed size doesn’t matter as much as total yield. For groundnut varieties harvested for human consumption (i.e., low oil, high O/L ratio) larger seed size is desired (Dwivedi et al., 2003).
Groundnuts are used in the food and confectionary industries, but is limited by its allergenic properties in both adults and children, and by concerns about aflatoxin contamination caused by Aspergillus niger and A. flavus), which is carcinogenic and act as an immunosuppressant in both animals and humans (Dwivedi et al., 2007).
Biotic Constraints
Disease Constraints
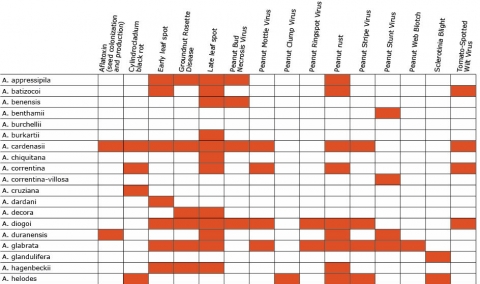
Biotic stresses include early leaf spot (Cercospora arachidicola), late leaf spot (Phaeoisariopsis personata), rust (Puccinia arachidis), groundnut mottle virus (Potyviridae), and groundnut rosette virus (Tombusviridae) (Okello et al., 2013; Pandey et al. 2012). (Table 2)
| Pathogen | Diseases |
| Fungi | Rust (Paccinia arachidis Speg.), early leaf spot (ELS) (Cercospora arachidicola Hori), and late leaf spot (LLS) [Phaeoisariopsis personata (Berk. and Curtis) Deighton] |
| Viruses | Groundnut rosette disease (GRD), peanut clump virus (PCV), peanut bud necrosis virus (PBNV), and tomato spotted wild virus (TSWV) |
| Bacteria | Bacterial wilt [Burkholderia solanacearum (E.F. Smoth) Yabuuchi et al.] |
| Nematodes | Meloidogyne, Scutellonema, Pratylenchus, Helicotylenchus, Aphelenchoides, Telotylenchus, and Paralongidorus species |
Pest Constraints
Important insect pests include aphids (Aphis craccivora), jassids (Amrasca devastans), leafminers (Aproarema modicella), termites (Isoptera), army worms (Spodoptera litura), and thrips (Frankliniella spp.) (Okello et al., 2013; Pandey et al., 2012) (Table 3). Thrips and aphids are more detrimental as vectors of viruses versus causing direct damage to the plants. The groundnut leaf miner (A. modicella) causes extensive defoliation in the major groundnut producing regions in Uganda (Mukankusi et al., 2000; Okello et al., 2013).
Because groundnut seed is sensitive to heat and high moisture, seed storage must be carefully managed at all stages of the seed production chain (Okello et al., 2013).
| Environment | Insect |
| Field | Leaf miner [Aproaerema modicella (Deventer)], army worm (Spedoptera litura Fab.), corn earworm (Helicoverpa armigera Hubner), lesser corn stock borer (Elasmopalpus lingosellus Zeller), southern corn rootworm (Diabrotica undecimpuctata howardi Barber), thrips (Frankinella and Scirtothrips species), jassids (Empoasca kerri Pruthi), aphids (Aphis craccivora Koch.), and termites (Microtermes and Odontotermes species) |
| Storage | Bruchid (Caryedon serratus Oliver), red flour beetle (Tribolioum castanem Herbst, rice moth (Corcyra caphalionica Sainton), and pod-sucking bug (Elasmolomus (Aphanus) sordidus Fab.) |
Resistance Traits
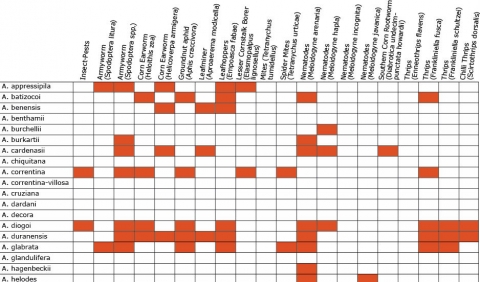
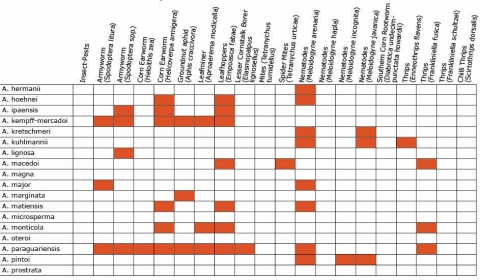
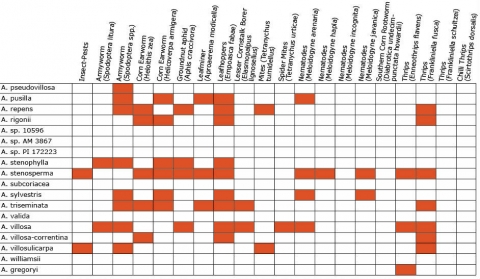
Wild Arachis species possess high levels of disease resistance and display a wide range of morphological variation (Dwivedi et al., 2007). e.g., A. diogoi has virus resistance genes that are not present in the cultivated gene pool (Table 4).
| Peanut accessions with beneficial traits reported | Type | |
| Trait | Cultivated Species | Wild Arachis species |
|---|---|---|
| Rust | 169 | 29 |
| Late leaf spot | 69 | 27 |
| Early leaf spot | 37 | 11 |
| Groundnut rosette virus | 116 | 12 |
| Nematode | 21 | – |
| Seed infection and/or aflatoxin production by Aspergillus flavus | 21 | 4 |
| Sclorotinia blight | 51 | – |
| Defoliator (Leaf miner and Spodoptera) | 9 | 28 |
| Aphid | 2 | Not evaluated |
| Drought | 40 | Not Evaluated |
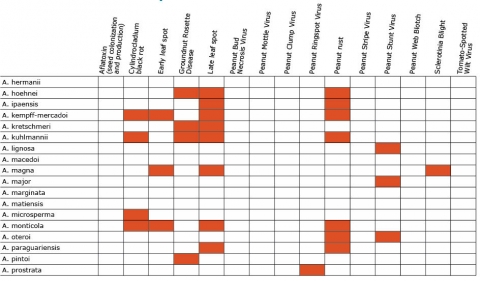
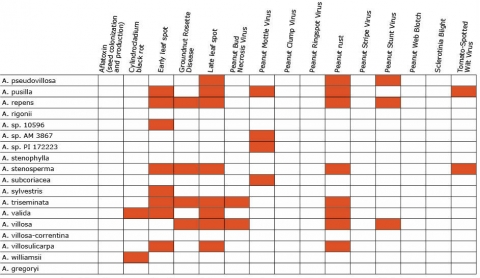
Resistance in Arachis Species (5 tables)
Figures 2 to 7 show Arachis species on the vertical axes and pests and diseases on the horizontal axes. Resistance is indicated by the red shaded boxes.
Breeding Properties
Breeding Goals
Improved yield – less vegetative biomass, shorter main stem, increased flower production (Dwivedi et al., 2003), high yielding potential, high quality, resistance to major pests and diseases (rosette and Cercospora leafspots, peanut bud necrosis, root rot), aflatoxin resistance, short to medium-term maturity periods and tolerance to drought, large seeds for confectionery purposes, high oil content for oil extraction (Okello et al., 2013).
Breeding CentersStalker et al., 2013.
The largest collection of groundnut germplasm is maintained in India at the International Crops Research Institute for the Semi-Arid Tropics (ICRISAT). This collection includes ~15,000 accessions of cultivated groundnut and ~500 accessions of 48 wild Arachis species representing all six botanical varieties: var. hypoagaea ( 45.8%), var. vulgaris (36.6%), var. fastigiata (15.7%), var. aequitoriana (0.10%), var. peruviana ( 1.7%), and var. hirsuta (0.13%).
Other major collections include the National Research Center for Groundnut in Junagadh, India (~8,000 accessions), the USDA NPGS, Griffin, GA, USA (~9,000 accessions). Large collections of wild Arachis species are also maintained at Texas A&M and North Carolina State University, Raleigh, NC, USA. The National Center of Genetic Resources (CENARGEN) in Brazil maintains over 1200 accessions of 81 species belonging to 9 sections.
Oil Crops Research Institute (OCRI) of the Chinese Academy of Agricultural Sciences (CAAS) (8083 accessions) and the Crops Research Institute of the Guangdong Academy of Agricultural Sciences (4210 accessions) in China
Instituto Nacional de Technologia Agropecuaria (INTA) and the Instituto de Botánica del Nordeste (IBONE) in Argentina
There are also two well-defined core and minicore collections representing the majority of variation present in the cultivated peanut germplasm.
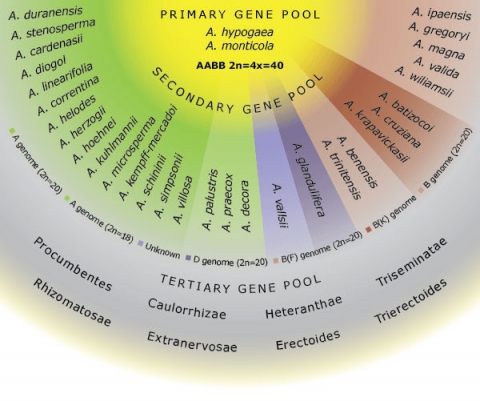
Germplasm Pools
The primary gene pool for groundnut consists of cultivated accessions (Arachis hypogaea) and the wild tetraploid species A.monticola. The secondary gene pool includes diploid species of the section Arachis that can be successfully crossed with cultivated groundnut. The tertiary gene pool includes species in sections other than Arachis that cannot be crossed with A. hypogaea using conventional crosses and are limited by both pre- and postzygotic hybridization barriers (Dwivedi et al., 2007) and (Fig. 8).
Introgressing Diversity from Wild Arachis Species
Wild species of Arachis have been successfully used as sources of resistance to pathogens (i.e., Sclerotinia blight, tomato spotted wilt virus, early and late leaf blights) and pests (root knot nematodes).
There are three main methods for the strategic incorporation of this diversity from wild species into cultivated groundnut (Dwivedi et al., 2007; Simpson, 2001).
Introgressing Diversity – Method 1
The first method starts with a cross between a diploid wild species and a tetraploid variety of cultivated groundnut (A. hypogaea), which produces a sterile triploid hybrid. The chromosomes of this sterile F1 hybrid are then doubled using colchicine to produce a hexaploid plant, which is then backcrossed with cultivated groundnut producing pentaploid progeny. These progeny are then self-pollinated for several generations usually resulting in aneuploid progeny, which over time lose chromosomes due to problems during meiosis (i.e., mispairing, lagging chromosomes) and eventually stabilize at the normal chromosome number of 40 (i.e., tetraploids) (Dwivedi et al., 2007; Holbrook and Stalker, 2003).
Introgressing Diversity – Method 2
For the second method, colchicine is used to double the chromosomes in two wild diploid species (i.e., AA and BB doubled to AAAA and BBBB) (Dwivedi et al., 2007). These autotetraploids are then crossed to produce an allotetraploid hybrid (i.e., AABB), which is then crossed and backcrossed with cultivated groundnut, A.hypogaea for multiple generations during which the breeder selects to recover useful agronomic traits from the cultivated parent while simultaneously selecting for exotic traits from the wild species.
For the third method two wild diploid species (i.e., AA and BB) are crossed and the sterile F1 hybrid (i.e., AB) is treated with colchicine to double the chromosomes to produce a fertile allotetraploid (i.e., synthetic amphidiploid, AABB) (Dwivedi et al., 2007). This allotetraploid is then backcrossed with cultivated groundnut A. hypogaea to produce tetraploid hybrids carrying agronomic traits from the cultivated parent and exotic traits from the wild species. Synthetic tetraploid varieties produced via this method are relatively normal and demonstrate normal meiosis, pollen fertility, and genetic recombination.
Introgressing Diversity – Method 3
Method 1 is the most useful for developing new varieties (reviewed by Dwivedi et al. 2003a and Holbrook and Stalker 2003). Method 2 is less useful for developing varieties in a breeding program because of problems with sterility in the autotetraploids (i.e., AAAA, BBBB) (Dwivedi et al., 2007). Method 3 is designed to recreate the events that originally gave rise to cultivated groundnut as described previously [i.e., hybridization of two 2x species (A genome from A. duranensis, B genome from A. ipaensis) followed by a spontaneous chromosome doubling of the sterile hybrid to form a fertile allotetraploid (AABB)]. This method has been used to introgress resistance genes from wild species into cultivated groundnut (Simpson and Starr 2001; Simpson et al. 2003). In this example Arachis cardenasii and A. diogoi were used as the A donors crossed with A. batizocoi, which was used as the B donor. The resulting allotetraploid was then crossed with cultivated groundnut (reviewed in Dwivedi et al., 2007).
Interspecific crosses are usually more successful when the species with the higher ploidy level (usually A. hypogaea) is used as the female parent when crossed with diploid wild Arachis species (i.e., Method 1). Greater success has also been observed when the annual species is used as the female parent and the perennial species (i.e., smaller stigmas) is used as the pollen parent. However, even when a cross successfully produces hybrid progeny, genetic recombination during meiosis in the hybrids is often restricted, and desired genes are not incorporated due to a lack of crossing over between the different genomes (Holbrook and Stalker, 2003). Because of these problems breeders must make multiple crosses using different parents to increase the probability of success.
Summary
Biparental crosses or more complex crosses are used to generate variability then pedigree and bulk-pedigree selection methods are used to identify and select superior genotypes. Single seed descent (with or without concurrent selection) is often used to increase the homozygosity of the breeding population during the early generations prior to selection (Dwivedi et al., 2003).
Typically breeders select for qualitative traits (such as disease/pest resistance) during the early generations F2-F5 followed by late generation testing for quantitative traits like yield and/or traits that are influenced by the environment (e.g., oil content, O/L ratio, etc.) (Dwivedi et al., 2003). Most groundnut breeding programs begin preliminary yield trials in the F5 or F6 generation, by which time the level of heterozygosity has been minimized through inbreeding and meaningful selection for complex quantitative traits can begin.
Recurrent selection is sometimes used to maintain genetic diversity in breeding populations (Dwivedi et al., 2003) though this method is limited by the presence of negative correlations between disease resistance and yield in some populations (Holbrook and Stalker, 2003).
Backcross breeding is commonly used for introgressing 1 or 2 genes into a superior genotype.
Outcomes
Artificial crosses are most successful when made during the early morning hours after sunrise when the pollen is mature and viable and the stigmas are receptive (Holbrook and Stalker, 2003). High humidity levels help ensure adhesion of pollen to the stigmatic surface so it is often helpful to spray down the greenhouse floors with water in the morning to increase the humidity levels especially on dry days (Holbrook and Stalker, 2003). Pod development in cultivated groundnut generally begins 16 to 17 days after pollination. In other species pod development is delayed until 23 to 25 days (Halward and Stalker 1987). Pegs of cultivated groundnut are relatively short and robust and do not break easily, but pegs of wild Arachis species may, since they may be 1 m or more in length and are fragile (Holbrook and Stalker, 2003), so it is preferable to use cultivated groundnut as the female parent whenever possible. Because only a small number of progeny (i.e., usually 1-2 seeds per pod) can be made per artificial cross it is important to make as many crosses as possible especially when making backcrosses to introgress traits from a wild Arachis species.
Outcrossing rates in groundnut are typically around 2% though rates near 8% have been identified (Coffelt, 1989; Knauft et al., 1992)
Because photoperiod and temperature greatly affect how growth is partitioned between above and below ground structures it is critical to make selections in an environment that is similar to your target environment. This is especially true in genotypes with large seeds. It is also important to select for resistance to pests and pathogens in an environment that has the same photoperiod conditions as your target environment because late maturing types are often more affected by variation in photoperiod (Dwivedi et al., 2003).
Limitations
Genetic enhancement of groundnut is limited due to the existence of unfavorable linkages between useful disease resistance genes and loci conferring undesirable pod and seed characteristics. A second limitation is due to the fact that most disease resistant accessions are later maturing types that are more sensitive to photoperiod and partition less to below ground pod development. A third limitation is the large genotype by environment interactions for economically important traits. A fourth limitation is the small number of progeny created by artificial crosses (Dwivedi et al., 2003).
Because of these limitations a varied approach to breeding groundnut is recommended (Dwivedi et al., 2003).
Breeding Strategies
| Trait | Conventional breeding | Marker-assisted selection | Wide crosses + marker-assisted backcross | Genetic transformation | Genetic basis |
|---|---|---|---|---|---|
| Maturity | X | n/a | n/a | n/a | Polygenic |
| Pod yield | X | n/a | n/a | n/a | Polygenic |
| Pod size and shape | X | n/a | n/a | n/a | Polygenic |
| 100-seed weight | X | n/a | n/a | n/a | Polygenic |
| Shelling outturn | X | n/a | n/a | n/a | Polygenic |
| Sound Mature seeds | X | n/a | n/a | n/a | Polygenic |
| Seed dormancy | X | n/a | n/a | n/a | Monogenic |
| Oleic/Linoleic fatty acid ratio | n/a | X | n/a | n/a | Oligogenic |
| Aflatoxin | n/a | n/a | n/a | X | Polygenic |
| Drought | n/a | X | n/a | X | Polygenic |
| Leaf miner | n/a | n/a | X | n/a | Unknown |
| Spodoptera | n/a | n/a | X | n/a | Unknown |
| Rust | X | n/a | n/a | n/a | Oligogenic |
| Early leaf spot | n/a | n/a | X | n/a | Polygenic |
| Late leaf spot | n/a | n/a | X | n/a | Polygenic |
| Bacterial wild | X | n/a | n/a | n/a | Oligogenic |
| Groundnut rosette disease | X | n/a | n/a | X | Mono- and diagenic |
| Peanut bud necrosis disease | n/a | n/a | n/a | X | Unknown |
Speed Breeding
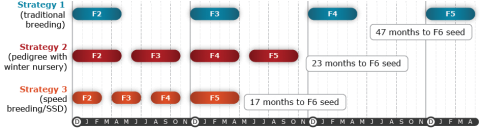
In temperate regions of the world, groundnut breeders are only able to grow 1 generation per year, thus it usually requires 10-15 years from the first cross to the release of a cultivar. When available the use of a winter nursery for increasing homozygosity levels via single seed descent can increase the total number of generations per year to 2, thereby reducing the time needed to develop a new cultivar. Recent efforts have demonstrated that the use of a controlled environment in which continuous light and optimal temperatures and humidity are maintained can be used to advance lines from the F2 to the F4 in a single calendar year, further reducing the time to release a cultivar down to 6-7 years. This concept of “speed breeding” allows the breeder to maximize the efficiency of the breeding program by decreasing the growing period to maturity by ~30% relative to the time needed in the field, thus increasing the number of generations that can be grown per calendar year to 3 (O’Connor et al. 2013).
- Strategy 1 (42 months): pedigree breeding with one summer generation per year
- Strategy 2 (23 months): pedigree breeding with two generations per year (i.e., 1 in the summer, 1 in a winter nursery)
- Strategy 3 (17 months): speed breeding/SSD (i.e., the F2-F4 generations grown in a controlled environment; 24 hour lights, optimal temperatures and humidity)
Calendar
In all 3 strategies, the F5 is grown in the field to maximize F5:6 seed numbers to facilitate preliminary yield trials in the F6.
Strategy 3 of speed breeding/SSD is best suited for programs using a backcrossing breeding strategy focused on incorporating a simply inherited trait controlled by one or two genes into a new variety. This system is also appropriate for the rapid development of RILs which are useful for genetic studies and molecular marker discovery.
Breeding Example
The following is an example of breeders introgressing alleles from Arachis cardenasii, a wild diploid species, into tetraploid groundnut.
Breeding objective: develop small-seeded, tetraploid (2n = 4x = 40) runner-type groundnut (Arachis hypogaea L. subsp. hypogaea var. hypogaea) germplasm lines that possess resistance to multiple diseases [e.g., early leaf spot (ELS), Cylindrocladium black rot (CBR), Sclerotinia blight (SB), and tomato spotted wilt (TSW)].
Registration of ‘Bailey’ Peanut
This is an example of breeders using simultaneous selection for resistance to multiple diseases using both early-generation testing for resistance combined with late-generation selection for improved pod and seed characteristics in superior families.
Breeding objective: develop a large-seeded virginia-type peanut (Arachis hypogaea L. subsp. hypogaea var. hypogaea) with partial resistance to five diseases that occur commonly in the Virginia-Carolina production area: early leaf spot (caused by Cercospora arachidicola Hori), late leaf spot [caused by Cercosporidium personatum (Berk. & M.A. Curtis) Deighton], Cylindrocladium black rot [caused by Cylindrocladium parasiticum Crous, M.J. Wingf. & Alfenas], Sclerotinia blight ( caused by Sclerotinia minor Jagger), and tomato spotted wilt (caused by Tomato spotted wilt tospovirus). This variety should possess characters that make it suitable for the confectionary market (i.e., sold to consumers as in-shell groundnut or as a shelled kernel).
The breeders used a modified version of pedigree selection that included several generations of advancement using single seed descent to develop a pureline cultivar named ‘Bailey’.
For the initial cross, NC 12C (a cultivar partially resistant to early leaf spot) was crossed with N96076L (a breeding line resistant to early leaf spot, CBR, and TSWV). Multiple F1 plants were then backcrossed as males to NC 12C to increase the chances of recovering an inbred line with suitable agronomic characteristics as NC 12C possessed more desirable traits relative to N96076L.
Why did the breeder backcross the F1s to the NC 12C parent? NC 12C was the superior plant.
Groundnut breeding is hampered by the limited genetic diversity in A. hypogeae and difficulty introgressing traits from wild Arachis species into cultivated varieties. The narrow genetic base in groundnut is attributed to multiple genetic bottlenecks that occurred as groundnut evolved into the current day species that it is, as well as, during domestication and as breeders work to develop modern-day cultivars. Consumers want a consistent commercial product so successful cultivars are often recycled back into a breeding program and used as the parents to develop the next round of cultivars.
References
Coffelt, T. A. 1989. Natural Crossing of Peanut in Virginia. Peanut Science 16(1): 46-48.
Dwivedi, S. L., J. H. Crouch, S. N. Nigam, M. E. Ferguson, and A. H. Paterson. 2003. Molecular breeding of groundnut for enhanced productivity and food security in the semi-arid tropics: opportunities and challenges. Advances in Agronomy 80:153-221.
Dwivedi, S. L., D. J. Bertioli, J. H. Crouch, J. F. Valls, H. D. Upadhyaya, A. Favero, M. Moretzsohn, and A. H. Paterson. 2007. Peanut. In: Oilseeds. Springer, Berlin Heidelberg, pp. 115-151.
Halward, T. M., and H. T. Stalker. 1987. Comparison of embryo development in wild and cultivated Arachis species. Ann. Bot. 59:9-14.
Holbrook, C. C., and H. T. Stalker. 2003. Peanut breeding and genetic resources. Plant breeding reviews 22:297-356.
Knauft, D. A., A. J. Chiyembekeza, and D. W. Gorbet. 1992. Possible Reproductive Factors Contributing to Outcrossing in Peanut (Arachis hypogaea L.). Peanut Science 19(1):29-31.
Kochert, G., H. T. Stalker, M. Ginenes, L. Galgaro, and K. Moore. 1996. RFLP and cytogenetic evidence for the progenitor species of allotetraploid cultivated peanut, Arachis hypogaea, (Leguminosae). Am. J. Bot. 83:1282-1291.
Krapovickas, A., and W. C. Gregory. 1994. Taxonomy of the genus Arachis (Leguminosae). Bonplandia 8:1-186.
O’Connor, D. J., G. C. Wright, M. J. Dieters, D. L. George, M. N. Hunter, J. R. Tatnell, and D. B. Fleischfresse. 2013. Development and Application of Speed Breeding Technologies in a Commercial Peanut Breeding Program. Peanut Science 40(2):107-114.
Okello, D. K., M. Biruma, and C. M. Deom. 2013. Overview of groundnuts research in Uganda: Past, present and future. African Journal of Biotechnology 9(39):6448-6459.
Pandey, M. K., E. Monyo, P. Ozias-Akins, X. Liang, P. Guimarães, S. N. Nigam, H. D. Upadhyaya, P. Janila, X. Zhang, B. Guo, D. R. Cook, D. J. Bertoli, R. Michelmore, and R.K. Varshney. 2012. Advances in Arachis genomics for peanut improvement. Biotechnology advances 30(3): 639-651.
Rami J-F., S.C.M. Leal-Bertioli, D. Fonceka, M.C. Moretzsohn, and D.J. Bertoli. 2014. Groundnut. In A. Pratap and J. Kumar (eds.), Alien Gene Transfer in Crop Plants, Volume 2:253-279.
Simpson C.E. 2001. Use of wild Arachis species/introgression of genes into A. hypogaea L. Peanut Sci 28:114-116.
Stalker, H. T., S. P. Tallury, P. Ozias-Akins, D. Bertioli, and S.C. Leal Bertioli. 2013. The value of diploid peanut relatives for breeding and genomics. Peanut Science 40(2):70-88.
Wynne, J. C., and W. C. Gregory. 1982. Peanut breeding. Advances in Agronomy 34:39-68.
Young, N.D., Weeden, N.F., and G. Kochert. 1996. Genome mapping in legumes (Fam. Fabaceae) in A.H. Paterson (ed) Genome Mapping in Plants. Landes, Co, USA pp. 211-227.

