Chapter 13: Sweetpotato Breeding
Jessica Barb and Anthony A. Mahama
Sweetpotato (Ipomoea batatas (L.) Lam., Convovulaceae) is a hexaploid species (2n = 6x = 90) that originated in both Central and South America. Recent evidence (Roullier et al. 2013) suggests that sweetpotato evolved from at least two autopolyploid events involving distinct populations of I. trifida or a now extinct species that was an ancestor to both I. batatas and I. trifida, which is similar to extant populations of wild tetraploid Ipomoea. The primary center of diversity for sweetpotato includes Central and South America (Huang and Sun, 2000; Zhang et al. 2000). Uganda in East Africa (Austin, 1987) (Yada et al 2010) and the region including Papua New Guinea (Fajardo et al. 2002) are both considered secondary centers of diversity.
- Become familiar with the basic biology and importance of sweetpotato
- Provide examples of breeding schemes used to develop cultivars of sweetpotato
Importance
Sweetpotato is a tremendously important low-input crop in Sub-Saharan Africa (SSA) that grows well without fertilizers and limited water and requires little care other than occasional hand weeding. In 2013 sweetpotato was the sixth most important food crop-based on total food crop production after cassava, maize, yams, rice, and sorghum, and 10th based on harvested land area (FAOSTAT).
Usage
Sweetpotato is grown for its enlarged storage roots and leaves that are harvested for human consumption (i.e., table stock), for animal feed, as a source of starch, and for industrial purposes. It is an important food security crop in African countries, especially in rural areas. Sweetpotato is a hardy crop that can provide reasonable yield in a variety of different environments and requires less external inputs (i.e., fertilizers, water) than most grain crops. Sweetpotato is a useful crop for subsistence purposes because it can be planted and harvested throughout the year in many locations, and a farmer can “store” roots in the ground and harvest the roots as needed. Roots are boiled, baked, fried, and included in a variety of dishes. Roots can also be dried and pulverized to make flour.
Sweetpotatoes are extremely variable in size, shape, color, moisture content, and carbohydrate content. Most consumers in SSA prefer the white-fleshed high dry matter types, but efforts are underway to develop orange-fleshed sweetpotato (OFSP) varieties that are both high in bioavailable beta-carotene and dry matter content to alleviate Vitamin A deficiency, especially in children. Sweetpotato leaves and petioles are good sources of protein, fiber, and minerals, especially K, P, Ca, Mg, Fe, Mn, and Cu, and when consumed could prevent malnutrition in developing countries. Purple fleshed types are also available that are usually high in dry matter and have a low level of sweetness.
Because sweetpotato can be planted throughout the year and there is a large range in maturity dates, farmers can manage the supply period and ensure continual yield, both for home consumption and for the local market. In most countries in SSA roots are available 4-8 months out of a year and in countries with two rainy seasons (i.e., Rwanda, Burundi, and Uganda) roots are available 11 months of the year. In most SSA countries except South Africa sweetpotato is grown primarily by smallholder producers who often plant a mix of different varieties in the same field, which is typically rainfed.
Important Diseases and Pests
Infected by Several Different Viruses
Sweetpotato is clonally propagated via vines, root slips (i.e., sprouts), or storage roots therefore, it is often infected by several different viruses. Durable resistance to viruses typically requires a combination of different traits that reduce the severity of symptoms, increase the tolerance of the variety in the field (i.e., decent yield despite the presence of the virus and visual symptoms), and allow the plant to recover from symptoms and revert to a virus free condition after being infected (Mwanga et al. 2013). Unfortunately degeneration of clean material often occurs very rapidly (i.e., within weeks to a year) (Gibson and Kreuze 2014) and yield losses are common.
Sweetpotato growing in humid, tropical, low- and mid-elevation regions of Eastern and Central Africa are mostly affected by Sweet potato chlorotic stunt virus (SPCSV) and Sweet potato virus disease (SPVD) which is caused by co-infection of Sweet potato chlorotic stunt virus (SPCSV) and Sweet potato feathery mottle virus (SPFMV). SPCSV is more detrimental than SPFMV and typically results in permanent symptoms and yield losses. SPCSV is transmitted by white flies. SPFMV is of less concern because infection of just this single virus only produces transient symptoms and very little loss in yield. This virus is transmitted by aphids. Resistance to SPCSV appears to be conferred by a recessive allele, which occurs in low frequency in the sweetpotato gene pool. However, this resistance still needs to be proven in regions where virus pressure is highest. SPVD (SPCSV + SPFMV) causes devastating yield losses in regions of high humidity though some varieties (e.g., NAPSPOT 11 and Tanzania) are reported to possess some levels of resistance. Other viruses that are less prevalent include: Sweet potato mild mottle virus (SPMMV), Sweet potato latent virus (SPLV), and Sweet potato virus G (SPVG).
Diseases and Pests in Africa
Sweetpotato growing in the humid, tropical highland regions of Eastern and Central Africa are affected by SPVD but are more affected by Alternaria stem blight which is the dominant disease of sweetpotato in this region. Other diseases that cause problems to a lesser extent throughout Africa include: scurf (caused by Monilochaetes infuscans), foot rot (caused by Plenodomus destruens), chlorotic leaf distortion (caused by Fusarium denticulatum), and Rhizopus soft rot (caused by Rhizopus stolonifer and R. arrhizus)
Two of the major pest groups that cause considerable damage to sweetpotato are plant-parasitic nematodes and weevils (Cylas spp.). Weevils are a major problem in drought-prone regions of Southern and Eastern Africa. Currently, there is no resistance to sweetpotato weevils.
Breeding Goals
Sweetpotato is grown for human consumption (i.e., table stock), processed starch, bioethanol, colorants/dyes, and for foliage for human and animal consumption. Examples of breeding goals include: resistance to sweetpotato virus disease (SPVD) and Alternaria stem blight, weevil resistance, improved yield, improved size, shape, and uniformity of roots, yield stability, high dry matter content, orange-flesh varieties with high nutritional value [i.e., beta-carotene content for combating vitamin A deficiency, improved chemical composition (i.e., starch, cellulose, sugars, protein content, carotenes, anthocyanins), improved micronutrient content (e.g., Zn and Fe)], extended harvest for subsistence cropping, drought tolerance, dense foliage with high protein content, improved palatability, and digestibility, vine survival and vigor after planting (especially during periods of drought), improved storage, resistance to skinning, and lower acrylamide potential.
Breeding Centers in Africa and Elsewhere
International Potato Center in Peru (CIP), Crops Research Institute (CSIR) in Ghana, Mozambique Institute of Agricultural Research (IIAM), National Crops Resources Research Institute (NaCRRI) in Uganda, North Carolina State University (NCSU) in the USA, Louisiana State University (LSU) in the USA, Agricultural Research Council (ARC) in South Africa, the Kenya Agricultural Research Institute (KARI), the Agricultural Research Institute (ARI) in Tanzania, the Zambia Agricultural Research Institute (ZARI), the Department for Agricultural Research Services (DARS) in Malawi, the Rwanda Agriculture Board (RAB), the National Root Crops Research Institute (NRCRI) in Nigeria, the Ethiopian Institute of Agricultural Research (EIAR) and the Environment and Agricultural Research Institute (INERA) in Burkina Faso.
Status of Sweetpotato in Africa (ca. 2015)
Constraints on Yield
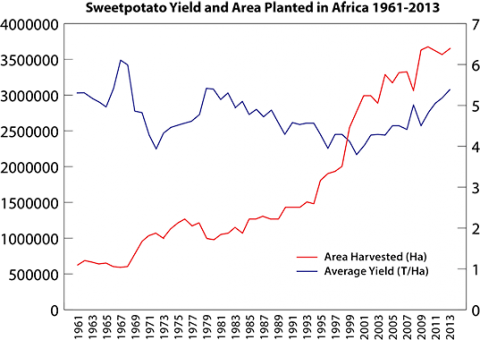
Although acreage in Africa planted with sweetpotatoes has steadily increased from the 1960’s, the average yield per hectare of sweetpotato has remained basically unchanged (Fig. 1) (FAOSTAT, 2014) due to five main constraints (Low et al. 2009):
- Farmers’ inability to acquire virus and disease free planting materials
- The lack of improved varieties
- The damage due to sweetpotato weevils
- Poor agronomic practices
- The lack of easily accessible markets
Average yield of sweetpotato in Africa is 5.6 tons ha-1, which is less than the average yield observed in the USA and China (24.5 and 22.4 tons ha-1, respectively), and far below the maximum achievable yield of 40-50 tons ha-1. Experiments conducted in East Africa suggest that yield could double if farmers had access to clean planting material of improved varieties, and the addition of improved crop and soil fertility management practices could more than triple the yield potential of sweetpotato in Africa (Gruneburg et al. 2004).
Yield and Acreage Over Time
The lack of improved varieties of sweetpotatoes is mostly the result of limited investment in sweetpotato breeding programs in Africa, though this trend is changing as government and NGO’s are now focusing more efforts towards the training and support of plant breeders working on secondary crops including sweetpotato. Additional effort is also now being focused on linking farmers with breeders to ensure that varieties produced by breeding programs are meeting the needs of farmers and consumers of sweetpotato.
Breeding Sweetpotato
Breeding Factors
The rate of progress that is achievable for a breeding program is dependent on the gene frequencies in the base population, the effectiveness of the breeding methods that are used, and the access the breeder has to field sites, greenhouses, lab equipment, and trained personnel needed to conduct a breeding program.
A breeder should consider the following when deciding on which breeding method to use:
- The germplasm that is available
- The inheritance of the target traits (if known)
- Biological constraints of the species (i.e., low seed set per plant, self-incompatibility, etc.)
Genetic improvement of sweetpotato is complicated by a number of factors:
- Self- and cross-incompatibility
- The highly heterozygous nature of individual clones
- The large number of chromosomes (2n = 6x = 90)
These factors contribute to a low correlation between parent performance and offspring performance. In general the success of a sweetpotato breeding program relies mostly on the ability to grow and evaluate a large number of clones/hybrid progeny in a selective environment that closely resembles the target environment. Thus the development of rapid and reliable screening methods is critical.
Hexaploid Nature of Sweetpotato
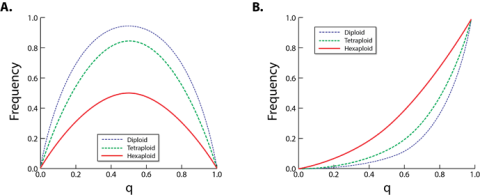
Cultivars of sweetpotato are phenotypically homogenous (because they are clonally reproduced) and genetically heterozygous (because they are self-incompatible outcrossers). Sweetpotato is a hexaploid with 2 non-homologous genomes (B1B1B2B2B2B2) with tetradisomic inheritance (Lebot 2010), so the genetics of simple traits is more complex with up to six alleles per locus. Furthermore, because sweetpotato is a hexaploid, heterozygous genotypes occur in much larger frequencies (Fig. 2) making heterosis more important for quantitatively inherited traits (e.g., yield, yield stability, vigor after planting, etc.).
The hexaploid nature of sweetpotato and the fact that this species is self-incompatible makes it extremely difficult to fix recessively inherited traits like resistance to certain viruses and some quality traits e.g., orange flesh) (Fig. 2) even if the frequency of these recessive alleles is greater than 70%.
Characteristics of Clonally Propagated Crops
Sweetpotato is an open-pollinated clonally propagated crop. Characteristics common to clonally propagated crops include:
- A strong positive relationship between productivity/vigor and level of heterozygosity
- Selfing reduces vigor due to inbreeding depression
- Vigor/heterozygosity can be fixed and maintained for the life of the clone
- Polyploids/aneuploids can be maintained via clonal propagation
- Often difficult to create a large quantity of clones from one plant, whereas it is relatively easy to produce a large amount of seed from most sexually propagated crops
- Clonal propagules are typically bulky and difficult to store
A basic breeding procedure for a clonal crop like sweetpotato includes the following steps:
- Define breeding objectives (i.e., yield stability, adaptation, taste, and pest and disease resistance)
- Assemble germplasm (i.e., local varieties, wild species, cultivars developed in other parts of the world, etc.) and establish a breeding nursery
- Develop segregating populations via hybridization (i.e., biparental cross, polycross nursery, or diallel crosses) and/or induced mutagenesis
- Evaluate and select superior clones
- Plot size, the number of replications, and the number of locations where clones are evaluated is increased after each round of selection in an attempt to reduce the variability due to the environment
- Select early for traits with high heritability, select later for traits with low heritability
- Name and release a cultivar and multiply and distribute clones
Basic Breeding Scheme for Sweetpotato
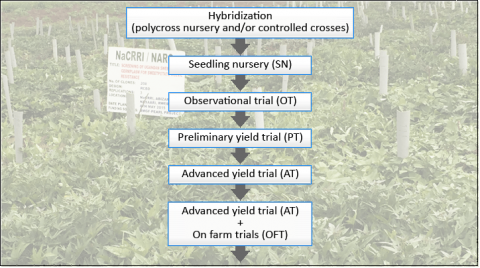
A basic breeding scheme for sweetpotato (Fig. 3) usually involves one (or more) cycles of hybridization to generate genetically variable progeny (i.e., true seeds) that are first evaluated in a seedling nursery. Superior genotypes are then clonally propagated and selection is conducted in replicated trials in multiple environments over multiple years (i.e., observational, preliminary, and advanced yield trials). Superior genotypes that are selected throughout the process are often recycled back into the system and used as parents for the next cycle which is the basis for recurrent selection.
Recurrent Selection
Recurrent selection is a common method for improving a breeding population that captures additive variance and is most effective for traits with moderate to high heritability. The basic scheme consists of three phases:
- Selection of genotypes and their hybridization in an insect-pollinated polycross nursery or using controlled crosses
- Evaluation of progeny
- Selection of superior progeny and the creation of a new polycross nursery or controlled crosses with or without the best parents from the initial polycross nursery in Step 1.
Recurrent selection is a proven method for increasing the frequency of desired alleles and for creating a genetically broad-based population especially when new germplasm (i.e., new parents/genotypes and/or superior progeny from previous cycles) is added to the polycross nursery/controlled crossing block after each cycle.
Recurrent selection allows for the rapid increase in the percentage of minor and recessive alleles, but it requires accurate screening techniques to be successful.
Accelerated Breeding Scheme (ABS)
Population Improvement and Variety Development
Many sweetpotato breeders are now using the Accelerated Breeding Scheme (ABS) for population improvement and for variety development (Table 1, below) (Gruneberg et al. 2009a, Gruneberg et al. 2015). The ABS reduces the total time needed to develop a new variety from 7-8 years to 4 years by compressing the early breeding stages into two years (i.e., 1 year of crosses/seedling multiplication + 3 to 4 years of evaluation in a single location vs. 1 year of crosses/seedling multiplication + only 1 year of evaluation in multiple locations). This method evaluates as many genotypes as possible in the first season of year 2 in 3-4 locations without replication. This allows the breeder to simultaneously select traits in multiple environments in a single year vs. growing clones in one location per year and sequentially selecting clones over several years. This shortens the time needed to develop a variety and conserves resources while still allowing the breeder to estimate the stability of the clones being tested. In sweetpotato, genotype x year variation (i.e., temporal variation) is typically less than genotype x location variation (i.e., spatial variation) (Gruneberg et al. 2004) so the focus is shifted to making selections in more environments in fewer years. For the second season of Year 2 a second replication and more locations are added.
G×E Interactions
High yield and stability in different environments is not well correlated in sweetpotato, suggesting that GXE interactions are important and yield needs to be evaluated under different conditions. For sweetpotato GXE interactions are often larger than or equal to the genetic variation for some traits (Gruneberg et al. 2009b) (Table 2, below); therefore, reducing replication to increase the number of locations can be beneficial. Because replication is eliminated in Season 1 of Year 2 an accurate understanding of the variability in the field is critical to ensure that the early evaluation stages are adequate for distinguishing superior genotypes from poor-performing genotypes. The inclusion of check varieties throughout the field (i.e., alpha lattice design) is suggested to account for microenvironments in a field that may bias the performance of individual clones.
The efficiency of the ABS can be increased by using a low input or high stress (i.e., drought) environment during Season 1 of Year 2 in addition to 2-3 normal input locations. Selection is conducted sequentially during Season 1 such that genotypes that don’t perform well in the low input/high-stress location are eliminated from the normal input locations before selections are made. This procedure is called “independent culling) (Gruneberg et al., 2009a) (Lebot 2010). This allows the breeder to select only genotypes that perform well both under normal and stressful environments in a single season.
In general breeding programs allocate >60% of the budget for replicated trials during the later stages when a few clones are evaluated in advanced yield trials in multiple locations. The accelerated breeding scheme shifts the emphasis of the budget to Year 2 to maximize the number of clones evaluated in multiple environments.
Accelerated Breeding Scheme
| Year | Season | Name of trial | # of clones/ genotypes | Plot type | # of clones/ genotypes | # of locations | # of reps/location | traits | Notes |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | Crossing block | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| 1 | 2 | Seeding nursery (SN) | 2000 | single hills (50cm x 50cm) | 1 | 1 | 1 | only highly heritable traits (i.e., disease-resistant, storage root color) | n/a |
| 2 | 1 | Observational trial (OT) | 2000 | single row plot (1m x 1m) | 3-4 | 3-4 | 1 | harvest index, storage root characteristics (i.e., dry matter content, protein, starch, sugars, provitamin A concentrations) | superior clones are often selected from OTs and used as parents for controlled crosses where the progeny are added to the next seeding nursery |
| 2 | 2 | Preliminary yield trial (PT) | 150-300 | 2 row plot | 30 plants (15 plants/row) | minimum of 3 | 2 | all traits | RCBD, 1 location can be used to create multiple environments if a treatment (i.e., fertilizer, irrigation) is applied |
| 3 | 1 | Advanced yield trait (AT) | 40 | 5 row plot | 75 plants (15 plants/row) | minimum of 4 | 2 | all traits | n/a |
| 3 | 2 | Advanced yield trial (AT) + On-farm trials (OFT) | 5-8 | 5 row plot | 75 plants (15 plants/row) | minimum of 4 plus 10 more on-farm locations (OFT) | n/a | all traits | field design/plot size determined by the farmer for OFTs |
| 4 | 1 | Multiplication | 1-3 | 1-3 ha fields | n/a | n/a | n/a | n/a | n/a |
Estimation of Variance Component
| n/a | Genotype
[latex]\sigma_G^2[/latex] |
Genotype by environment
[latex]\sigma_{G \times E}^2[/latex] |
Error
[latex]\sigma_E^2[/latex] |
Ratio
[latex]\sigma_G^2:\sigma_{G \times E}^2:\sigma_E^2[/latex] |
|---|---|---|---|---|
| Yield (t2/ha) | 36.2
(20.6 – 43.6) |
39.4
(33.8 – 46.9) |
64.2
(60.0 – 68.9) |
1 : 1.1 : 1.8 |
| Dry matter (%2) | 14.8
(13.3 – 16.6) |
5.7
(5.0 – 6.5) |
5.7
(5.3 – 6.1) |
1 : 0.4 : 0.4 |
| Protein (%2 DM+) | 0.21
(0.16 – 0.30) |
0.67
(0.59 – 0.78) |
0.73
(0.68 – 0.79) |
1 : 3.2 : 3.5 |
| Starch (%2 DM+) | 21.5
(19.3 – 24.2) |
3.2
(2.3 – 4.9) |
16.3
(15.2 – 17.5) |
1 : 0.2: 0.8 |
| Sucrose (%2 DM+) | 5.6
(4.9-6.5) |
2.1
(1.6 – 2.8) |
7.4
(6.9 – 7.9) |
1 : 0.4 : 1.3 |
| Total sugar (%DM+) | 17.0
(15.2 – 19.2) |
6.0
(5.2 – 7.1) |
9.0
(8.4-9.7) |
1 : 0.4 : 0.5 |
| Carotene (ppm2 DM+) | 6327
(5681-7091) |
2462
(2224 – 2740) |
1421
(1323 – 1529) |
1 : 0.4 : 0.2 |
| Calcium (ppm2 DM+) | 74001
(61485-90791) |
95.990
(82657 – 112849) |
157303
(147021-168708) |
1 : 1.3 : 2.1 |
| Magnesium (ppm2 DM+) | 143005
(12351 – 16764) |
9880
(8360 – 11858) |
17638
(16451 – 18960) |
1 : 0.05 : 0.1 |
| Iron (ppm2 DM+) | 2.33
(1.92 -2.87) |
3.46
(3.0 – 3.97) |
3.85
(3.59 – 3.97) |
1 : 1.7 : 2.2 |
| Zinc (ppm2 DM+) | 0.8
(0.62- 0.97) |
1.37
(1.20 – 1.59) |
1.72
(1.60- 1.85) |
1 : 1.7 : 2.2 |
Establish a Crossing Block
YEAR 1 (SEASON 1) – ESTABLISH A CROSSING BLOCK AND/OR MAKE CONTROLLED CROSSES AND HARVEST TRUE (HYBRID) SEED
Sweetpotato breeding has traditionally involved making crosses between complementary parents using controlled crosses or a polycross nursery with multiple parents.
The major goal of a crossing block or controlled crosses is the improvement of the overall population mean from one cycle to the next.
The ability to generate genetic variation is easy in sweetpotato due to the high heterozygosity of individual clones/parents.
The creation of genetic variation via hybridization is easy in sweetpotato because this species is generally self-incompatible so seeds do not develop from self-fertilization.
If a breeder has limited information about the value/usefulness of individual crosses (i.e., from prior test crosses or data from previous years) then the goal should be to maximize the number of parental combinations.
A minimum of 15 parents should be used in a polycross nursery or for controlled crosses. Most breeding programs use 20-30 parents – though larger breeding programs (e.g., CIP) are now using 150 or more different parents that are separated into different gene pools (i.e., heterotic groups).
YEAR 1 (SEASON 2) – SEEDLING NURSERY (SN)
Hybrid seed from the crossing block or controlled crosses is harvested and planted in 0.5 x 0.5 m plots (depends on quantity of seed available and the size of the breeding program). Selection during this step is usually limited to natural selection for tolerance and resistance to pathogens and pests. Artificial selection by breeders is typically avoided during the “true seed plant stage” because seed plants often grow differently than plants grown from vegetative cuttings and because this stage is often grown in an artificial environment (e.g., shade or glass house in pots) that is not representative of the field environment where the clones will eventually be grown. Stem cuttings are harvested after 10 weeks.
YEAR 2 (SEASON 1) – OBSERVATIONAL TRIAL (OT)/HILL TRIAL (HT)
Hybrid seed from the crossing block or controlled crosses is harvested and planted in 0.5 x 0.5 m plots (depends on quantity of seed available, size of breeding program). Selection during this step is usually limited to natural selection for tolerance and resistance to pathogens and pests. Artificial selection by breeders is typically avoided during the “true seed plant stage” because seed plants often grow differently than plants grown from vegetative cuttings and because this stage is often grown in an artificial environment (e.g., shade or glass house in pots) that is not representative of the field environment where the clones will eventually be grown. Stem cuttings are harvested after 10 weeks.
YEAR 2 (SEASON 2) – PRELIMINARY YIELD TRIAL (PT)
Larger 2 row plots (30 plants, 15 plants/row) are planted at a minimum of 3 locations. A second rep is added at each location. Selection for lower heritability traits (e.g., yield, biomass) begins at this stage. A breeder can begin to assess yield stability at this step but this requires 3 or more environments with the best results achieved when more than 6 environments are included (Gruneberg et al. 2009a).
YEAR 3 (SEASON 1) – ADVANCED YIELD TRIAL (AT)
Larger 5 row plots (75 plants, 15 plants/row) are planted at a minimum of 4 locations. Selection for lower heritability traits (e.g., yield, biomass) continues.
YEAR 3 (SEASON 2) – ADVANCED YIELD TRIAL (AT) + ON FARM TRIALS (OFT)
On farm trials are added where the field design, plot size, and inputs are determined by the individual farmers. Farmers are asked to rate the clones on a common set 5-6 traits plus any additional traits that are important to them.
YEAR 4 (SEASON 1 AND 2) – MULTIPLICATION AND DISTRIBUTION OF VARIETIES
Superior genotypes are multiplied and distributed to farmers
Trial Conditions
A breeder is strongly encouraged to plant check varieties, guard rows, and spreader/susceptible rows and to use an appropriate field design for each location. It is best to harvest cuttings from plants that are ten weeks old, so it is helpful to plant a multiplication plot for each clone about 2.5 months before a trial is planted. These plots should be maintained separately and managed (e.g., weeded, fertilized, irrigated) to promote vegetative growth. A single vine cutting usually produces five vine cuttings. Storage root sprouts can also be used (i.e., an average-sized storage root yields about 20 vine cuttings). A breeder should use tip cuttings of the same length and vigor when possible. If uniform cuttings are not available divide the cuttings up by replication (i.e., best in rep 1, second-best in rep 2, etc.). All field management (e.g., weeding, fertilizer, irrigation, etc.) should be applied uniformly across all reps. If possible each task should be divided so each worker completes an entire rep. For example, if planting an entire field takes 3 people, 1 worker should plant rep 1, a second worker would plant rep 2, and a third worker would plant rep 3, so differences in planting technique can be partitioned out as a rep effect when the data is analyzed. The same procedure should be applied for data/note-taking. If multiple people are making evaluations each individual should be assigned to evaluate specific reps and workers should not collect data for others.
It is nearly impossible to simultaneously select the top genotypes for every trait without growing out an impracticably large population because the number of genotypes to be screened increases exponentially as the number of traits increases. For example, if a breeder wanted to select the top 10 clones from among 100 genotypes for 10 traits then 100^10 genotypes need to be grown and evaluated. Thus a breeder usually has to compromise and select 3-5 quality traits at a time while simultaneously maintaining sufficient genetic variation for yield, yield stability, and adaptability.
ABS – Mulitple Concurrent Cycles
Breeders will usually have 3-5 different cycles of an ABS at various stages (SN, HT/OT, PT, AT) running at the same time (see Fig. 4). This allows a breeder to use advanced clones from later stages of one cycle to create new polycross nurseries/controlled crosses.

Sources of Parents/Germplasm
Institutions that maintain and distribute Ipomoea germplasm include: CIP (Centro Internacional de la Papa) (> 4,200 accessions), USDA/ARS National Genetic Resources Information Network (>900 accessions), NaCRRI National Crops Resources Research Namulonge/Uganda, and IITA (International Institute of Tropical Agriculture), in Nigeria.
Though some interspecific hybrid combinations can be made especially using species within the Batatas section, wild species are typically not used in most breeding programs.
Polycross Nursery vs. Controlled Crosses
Ensuring Specific Cross Combinations
Often a combination of open-pollination (i.e., polycross nursery) and hand-pollination (i.e., controlled crosses) is used to ensure that specific cross combinations are made. Some of the advantages and disadvantages of using a polycross nursery vs. controlled crosses are shown in Table 3.
| Polycross nursery | Controlled crosses | ||
| Advantages | Disadvantages | Advantages | Disadvantages |
| Requires less labor |
Only the female parent is known so genetic advancement is based on only ½ of the genetic variation that is available |
Both parents are known so genetic advancement is based on all of the genetic variation that is available | Requires more labor to make crosses by hand |
| Makes more seed/more cross combinations | Unbalanced contribution from some clones (poor seed set, low pollen production, limited and/or asynchronous flowering) | Superior combinations are tracked and can be recreated | Less seed/less combination are created |
Polycross nurseries are theoretically less efficient because genetic advancement is halved because parental control is based only on the female parent while parental control for controlled crosses is based on a control factor of 1 because both parents are known.
GS = (c)(i)VA / σP
GS is the expected gain or predicted genetic advance from selection
c = parental control factor, ½ if only the female parent is known,1 if both parents are known
i = the selection intensity, a constant based on the percent selected and obtained from statistical tables
VA = additive genetic variance
σP= phenotypic standard deviation
Note: the phenotypic values must exhibit a normal, or bell-curve, distribution for GS to be valid.
Using Different Methods
To better understand the impact of using different methods to generate progeny, CIP breeders and collaborators compared the population means (i.e., unselected and after one cycle of selection) of sweetpotato progeny created by 3 different pollination designs (I, II, and III) to determine which method of generating progeny produced the best results. For this experiment the breeders used the same 22 clones but crossed them in different ways: (I) using an open-pollinated polycross nursery, (II) using a partial diallel design where 4 of the best clones were crossed by hand to each other and to the rest of the clones (4 x 22), and (III) using a factorial design where the best 5 clones were crossed by hand to rest of the remaining clones (5 x 17) (“the best by the rest”). The progeny of each of these designs were planted in unreplicated plots in a single location and the population mean was determined for each = average performance of the unselected population. The breeders then selected the best 100 genotypes from each population and averaged their performance as a measure of the achieved increase in root yield after one cycle of selection. The standardized response to selection (R) was then calculated for each method by comparing the unselected mean to the mean of the 100 selected genotypes. The results for this experiment showed that the average root yield of the progeny created by method (II) (18.4 t/ha) and the average root yield of the 100 selected genotypes (23.5 t/ha) after one round of selection were both higher than the unselected and selected progeny generated by the other methods (I and III) (Table 4). These results indicated that for this scenario, controlled crosses made by a breeder guided by a partial diallel design (II) produced better progeny and more genetic advancement per cycle than an open-pollinated polycross nursery (I). The factorial design (III) was the least successful method though it only differed from the partial diallel design in that it did not include progeny from the intermating of the top 5 clones. This suggests that much of the success of the partial diallel design may be attributed to the successful performance of the progeny created by crossing the top 5 clones.
Case Study: Comparing the Use of a Polycross Nursery vs. Controlled Crosses
Considering Other Factors
| Design | Pollination design | Population mean of progeny before selection (t/ha) | Population mean of progeny before selection (t/ha) | Standardized R |
|---|---|---|---|---|
| I | Open pollinated polycross nursery (22) | Open-pollinated polycross nursery (22) | 21.2 | 1.35 |
| II | Partial diallel (4 X 22) | Partial diallel (4 X 22) | 23.5 | 0.904 |
| III | Factorial (5 X 17) | Factorial (5 X 17) | 17.4 | 0.715 |
Despite the gains made using controlled crosses vs. a polycross nursery in this particular scenario a breeder must also consider other factors as well when deciding which method to use for their program. For example, not all programs have enough staff to make all of the hand pollinations necessary for a partial diallel design so a polycross nursery may be a better option. If a breeder does choose to use an insect-pollinated polycross nursery it is important that all clones be equally represented so experimental design and replication are crucial. If a polycross nursery contains ten or fewer clones a Latin square design is recommended (Table 5). For a large number of clones a randomized complete block design with replication is recommended (Table 5).
Hybridization Using a Polycross Nursery
| Latin square – n × n array, each genotype occurs exactly once in each row and exactly once in each column | Randomized complete block design | ||||||||||
| A | B | F | C | E | D | A | B | C | C | A | E |
| B | C | A | D | F | E | D | E | F | B | H | F |
| C | D | B | E | A | F | G | H | I | D | I | G |
| D | E | C | F | B | A | F | A | G | I | A | B |
| E | F | D | A | C | B | B | D | H | G | H | F |
| F | A | E | B | D | C | E | C | I | D | C | E |
- Planted in isolation
- Natural insect cross-pollination & artificial crosses to ensure random pollination
- Plant extra reps of less vigorous clones or clones that don’t flower well
- Stagger planting times so that all clones flower at the same time
Crossing Block/Controlled Crosses
- Flowering is best under short day conditions and begins about 1.5 months after the crossing block is planted/established and continues for ~3-5 months
- It is advisable to plant extra replications of clones that do not flower well
- Clones that don’t flower well can also be grafted onto I. setosa, or I. nil, or on a genotype that flowers prolifically to promote flowering
- Clones that flower later than the other clones should be planted earlier to ensure that all clones have an equal chance of contributing to the gene pool of the progeny
- Seed harvest begins about 2 months after the crossing block is established and continues for up to 4 months
- Plant 2 vines per 1×1 m plot, 5 randomized plots of each clone
- Stake/trellis plants and erect a windbreak if needed to protect the crossing block from high winds
- Label each stake with the clone number to make identification easier
- Avoid using a high N fertilizer to promote flowering
- Monitor plants for insect and disease problems, but avoid using pesticides that may injure bees, and other pollinating insects
- Most clones of sweetpotato are self-incompatible and do not produce selfed seed
- Each flower opens early in the day just after sunrise and lasts for only a few hours before fading around noon
- Each pistil contains 1 superior ovary with two carpels and each carpel has two locules that contain 1 or 2 ovules, so a single capsule can produce 1 to 4 seeds
- Flowers that are hand-pollinated usually produce 1 or 2 seeds and capsules that are open-pollinated produce 2 to 3 seeds.
Seed Harvest
Seeds mature 4 to 6 weeks after pollination. The capsule and pedicel will both turn brown and dry and begin to shrivel when the seed is ready to harvest. Capsules that are left too long will dehisce (split open) so care must be taken to harvest seeds before they are lost.
Seed Scarification
Seeds can be hand-scarified by scratching a small notch in each seed coat with a sharp needle or a small, 3-cornered file or acid scarified with concentrated sulfuric acid for 40 minutes followed by a 5-10 minute rinse under running water.
Polycross Nursery vs. Controlled Crosses
Cross Incompatibility
Self-fertilization is rare because sweetpotato possesses a homomorphic, sporophytic type of self-incompatibility that is not affected by environment, chemical treatment, and cannot be overcome using bud pollination. This system is likely controlled by a single S-locus with multiple alleles with a dominant-recessive relationship. Heterostyly also occurs in sweetpotato; however, it does not appear to affect fertility.
Cross incompatibility among different varieties can limit recombination and seed production and hinders targeted breeding especially when the parents with desirable traits of interest, such as disease resistance, drought tolerance, enhanced levels of protein, vitamins, macro- and micro-nutrients and dry matter are closely related and belong to the same incompatibility group. As a result breeders must maintain large populations that contain non-related accessions with complementary traits. Three types of cross compatibility (Table 6) exist depending on the success or failure of reciprocal crosses: reciprocal fertility occurs when fertility is present in both directions, reciprocal incompatibility occurs when incompatibility occurs in both directions, and unilateral fertility/incompatibility occurs when fertility occurs only when a genotype is used as the female but not when used as a male and vice versa.
| Type of cross-compatibility | Expected outcome; A and B are different genotypes |
|---|---|
| Reciprocal fertility | A x B and B x A both produce seed |
| Reciprocal incompatibility | A x B and B x A both do not produce seeds |
| Unilateral fertility/incompatibility | A x B produces seed but B x A does not OR
B X A produces seed but A x B does not |
Incompatibility and Sterility
Incompatibility and sterility are often used interchangeably although this is done incorrectly. Sterility is the failure of reproduction due to the failure of a plant to produce viable gametes and incompatibility is the failure of viable gametes to fertilize one another. Sterility or reduced fertility in sweetpotato is not uncommon as aneuploidy due to multivalent formation among non-homologous chromosome pairs can often lead to an unbalanced number of chromosomes in the gametes. Sterility is also caused by gene action.
Participatory Plant Breeding (PPB)
Women are responsible for most of the labor when it comes to growing sweetpotato. Although men are usually involved in land preparation, especially when land needs to be cleared or soils are heavy, and in marketing, particularly where sweetpotato is a significant cash crop. Men also significantly contribute to weeding and harvesting, particularly when sweetpotato is intercropped.
Traits that are important to farmers (e.g., piecemeal harvest for subsistence farming and resistance to regionally important abiotic and biotic stresses, etc.) are difficult to select for outside of the target environment(s), therefore, it is helpful to cooperate with local farmers/growers to gain access to additional testing locations and to gain insight into farmer/grower preferences. Participatory plant breeding practices are beneficial especially during the earlier steps when genetic diversity is high and most traits (e.g., quality characteristics, disease resistance, plant architecture, etc.) are visually evaluated. PPB practices are less useful at the later stages of selection because those traits that are usually selected based on visual evaluations are typically highly heritable and are already fixed in the population. The involvement of farmers and consumers also helps facilitate the rapid adoption of new varieties and lessens the chance that superior varieties are rejected because they fail to meet the expectations of the farmers. Regional and local preferences for flesh color, dry matter content, and texture, as well as adaptability to the local environment are critical traits in sweetpotato and should be considered during the early stages of selection.
CIP’s Convergent-Divergent Scheme
CIP breeders are currently using a convergent-divergent approach that is designed to meet the regional needs of growers by developing widely adapted cultivars and promoting collaboration among breeders and consumers (Lebot 2010). Due to the difficulty of breeding for adaptation across multiple agro-ecological zones in Africa a program must be conducted in a decentralized way. Therefore programs in different countries can develop cultivars that work best for their region. This method starts with a diverse base population composed of genotypes from a wide range of sources. This base population is evaluated in a single centralized location and superior genotypes are intercrossed. Seed from this cycle is collected and then sent to collaborators in different regions where it is evaluated and intercrossed with elite germplasm that is adapted for each individual location. This scheme introduces new germplasm while also including elite germplasm carrying alleles for traits that are specific to a region (e.g., disease and pest resistance endemic to that location, a particular taste profile, plant architecture that fits a cultivation technique used in that region, etc.). By allowing breeders to introduce alleles specific to their location and then test the progeny locally GXE interactions are better accounted for. Selection in the centralized location is focused on maintaining high genetic diversity while also introducing desired traits, while selection at the local level focuses more on selecting varieties that will grow well in a specific environment.
Exploiting Heterosis
Creating Distinct Germplasm Pools
Exploiting Heterosis Using 2 Heterotic Gene Pools
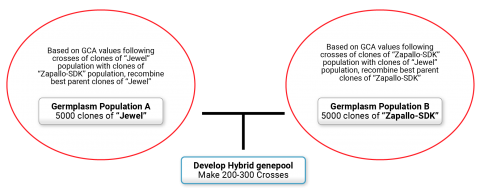
Breeders are now exploring the idea of exploiting heterosis in sweetpotato by creating two genetically distinct germplasm pools and selecting and recombining within each pool based on the general combining ability (GCA) of clones that are crossed with the other germplasm pool. Work at the international Potato Center (CIP) is focused on creating high yielding orange-fleshed sweetpotato varieties using this approach. For this experiment, two populations were created: the Jewel population and the Zapallo population. The Jewel population consists of orange-fleshed clones released prior to 2004 and includes mostly North American varieties including Jewel and Resisto. The Zapallo population was created using a factorial crossing design using 8 male parents (Jonathan, Zapallo, Huambachero, Tanzania, Yurimaguas, Wagabolige, Xushu18, and Ninshu1) and 200 orange fleshed females. More than 200 families were then produced from cross combinations involving 49 clones from the Jewel population crossed with 31 clones from the Zapallo population (Fig. 5). Mid-parent (MP) heterosis values for dry root yield were generally low to moderate, but some individual cross combinations/families had MP heterosis values. Root yield of the best clone within each family was typically double the average root yield of the parents, with some clones producing 5 times the average yield of the parents, thus indicating success of the approach.
Crossing Scheme and Heterosis Values
Choosing Parents
Example of Parent Selection
Choosing Parents for a Crossing Block/Controlled Crosses
A breeder will often use morphological data, coancestry/pedigree information, and molecular marker data to choose parental combinations. By using a combination of analyses a breeder can select parents that are phenotypically superior, and genetically distant from one another to limit inbreeding depression and possible cross-compatibility issues. Molecular markers are also useful for identifying duplicate clones or instances when genetically different clones are distributed under the same name.
Example of Parent Selection Based on Molecular Markers vs. Morphological Markers
Clones were characterized using both molecular markers and agronomic characters to identify phenotypically superior yet distantly related clones (Dai-fu et al. 2009). Fifteen clones (Table 7) and 60 open-pollinated offspring per clone were planted in a common garden and phenotyped for 22 traits. The 15 parental clones were genotyped with ISSR and RAPD markers. The dendrograms (Figs. 6 and 7) based on the morphological traits and the molecular markers are shown below along with Table 8, which summarizes the different groupings based on the morphological and molecular marker data.
Trial Materials and Parental Dendrograms
Choosing Parents for a Crossing Block/Controlled Crosses
| Code | Variety | Female1) | Male | Source or distribution location |
|---|---|---|---|---|
| 1 | Centennial | Unit 1 Porto Rico | Pelican Processor | Louisiana State University, USA |
| 2 | Beijing 553 | Okinawa100 OP | The Experimental Station of Huabei, China | n/a |
| 3 | Beauregard | L78-21 OP | Louisianna State University, USA | n/a |
| 4 | Chaun 8129-4 | Jiangjin Wujianshao | Neiyuan | The Crop Institute, Sichuan Academy of Agricultural Sciences, China |
| 5 | Guangshu 88-70 | Zhan75-57 OP | The Crop Institute, Guangdong Academy of Agricultural Sciences, China | n/a |
| 6 | Jianshui Huangxin | Landrace | Jianshui County, Yunnan Province, China | n/a |
| 7 | Longshju 1 | Yanfen 1 | Longyan 7-3 | Agricultural Science Institute of Longyan, Fuijian, China |
| 8 | Sushu 6 | Peng S1-12-48 | AIS 0122-2 | Xuzhou Sweetpotato Research Centre, Jiangsu |
| 9 | Tainung 69 | Group crossing | Jiayi Agricultural Experimental Station, Taiwan of China | n/a |
| 10 | Xiangshu 15 | Kyushu 5 | Xiangshu 6 | The Crop Institute, Hunan Academy of Agricultural Sciences, China |
| 11 | Xu 22-5 | LO323 | AIS 0122-2 | Xuzhou Sweetpotato Research Centre, Jiangsu, China |
| 12 | Xu 34 | Huabei 166 | Qunli 2 | Xuzhou Sweetpotato Research Centre, Jiangsu, China |
| 13 | Xushu 23 | P616-23 | Yanshu 27 | Xuzhou Sweetpotato Research Centre, Jiangsu, China |
| 14 | Yanshu 27 | 77183 | Centennial | Agricultural Science Institute of Yantai, Shandong, China |
| 15 | Yanshu 5 | Yanchi Red | Yan 94-1 | Agricultural Science Institute |
1) OP, open pollination
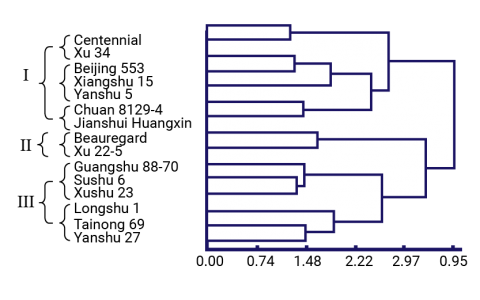
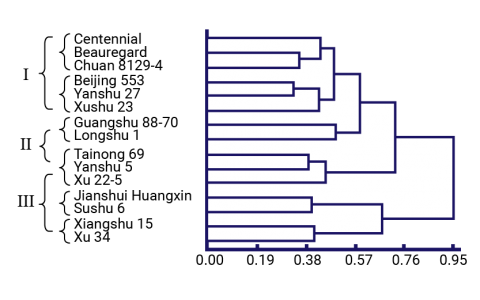
Summary of Groups of Clones
Choosing Parents for a Crossing Block/Controlled Crosses
| Clone | Phenotype Group | Genotype Group | |
|---|---|---|---|
| Centennial | 1 | 1 |  Choose 1 from this group of 3 Choose 1 from this group of 3 |
| Beijing 553 | 1 | 1 | |
| Chuan 8129-4 | 1 | 1 | |
| Yanshu 5 | 1 | 2 |  Choose this one Choose this one |
| Xu 34 | 1 | 3 |  Choose 1 from this group of 3 Choose 1 from this group of 3 |
| Xiangshu 15 | 1 | 3 | |
| Jianshui Huangxin | 1 | 3 | |
| Baeuregard | 2 | 1 |  Choose this 1 Choose this 1 |
| Xu 22-5 | 2 | 2 |  Choose this 1 Choose this 1 |
| Xushu 23 | 3 | 1 |  Choose 1 from this group of 4 Choose 1 from this group of 4 |
| Yanshu 27 | 3 | 1 | |
| Guangshu 88-70 | 3 | 1 | |
| Longshu 1 | 3 | 1 | |
| Tainong 69 | 3 | 2 |  Choose this 1 Choose this 1 |
| Sushu 6 | 3 | 3 |  Choose this 1 Choose this 1 |
A breeder can use this information to select clones that are phenotypically similar (i.e., same phenotype group) but genetically different (i.e., different genotype group) thereby reducing the number of parental clones from 15 to 8 without significantly reducing the genetic variation that is available.
Combining Ability of Individual Clones
Choosing Parents for a Crossing Block/Controlled Crosses
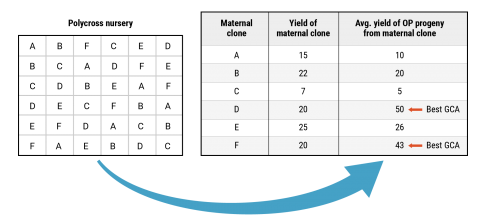
When considering which parents to choose for a polycross nursery or controlled crosses it is also important to consider the combining ability of individual clones. The combining ability of a clone can be estimated by comparing the performance of a clone versus the average performance of open-pollinated progeny harvested from that clone when it is grown in a polycross nursery or other population of mixed genotypes (i.e., seedling nursery, yield trial, etc.), for example Figure 10. If the average performance of the progeny is greater than the performance of the maternal clone then a breeder can assume that a particular clone has good combining ability when crossed with other clones.
Parent Selection
Important to choose parents that possess superior qualities and parents that combine with other clones (superior GCA)
References
Austin, D.F. 1987. The taxonomy, evolution and genetic diversity of sweet potatoes and related wild species. In: Exploration, Maintenance, Utilization of Sweet Potato Genetic Resources. Proceedings of the 1st Planning Conference, Lima, Peru, International Potato Center (CIP), pp. 2759.
Dai-fu et al. 2009. Selection of parents for breeding edible varieties of sweetpotato with high carotene content. Agricultural Sciences in China 2009, 8(10): 1166-1173.
Fajardo D.D., D.R. La Bonte, and R.L. Jarret. 2002. Identifying and selecting genetic diversity in Papua New Guinea sweet potato Ipomoea batatas (L.). Lam. germplasm collected as botanical seed. Genet. Resour. Crop Evol. 49: 463-470.
Gallais, A. 2003. Quantitative genetics and selection theory in autopolyploid plants. France, INRA.
Gibson, R. W., and J. F. Kreuze. “Degeneration in sweetpotato due to viruses, virus‐cleaned planting material and reversion: a review.” Plant Pathology 64, no. 1 (2015): 1-15.
Grüneberg, W.J., E. Abidin, P. Ndolo, C.A. Pereira, and M. Hermann. 2004. Variance component estimations and allocation of resources for breeding sweet potato under East African conditions. Plant Breeding 123: 311-315.
Grüneberg, W.J., R. Mwanga, M. Andrade, and J. Espinoza. 2009a. Selection methods Part 5: Breeding clonally propagated crops. In: Plant Breeding and Farmer Participation (S. Ceccarelli, E.P. Guimarães, and E. Weltzien, eds.). FAO, Rome, pp. 275-322.
Grüneberg, W.J., Mwanga, R., Andrade, M. and Dapaah, H. 2009b. Sweetpotato breeding. In: Andrade, M.,
Barker, I., Cole, D., Dapaah, H., Elliott, H., Fuentes, S., Grüneberg, W.J., Kapinga, K., Kroschel, J.,
Labarta, R., Lemaga, B., Loechl, C., Low, J., Lynam, J., Mwanga, R., Ortiz, O., Oswald, A. and
Thiele, G. Unleashing the Potential of Sweetpotato in Sub-Saharan Africa: Current Challenges and
Way Forward. Working Paper 2009-1. International Potato Center (CIP), Lima, pp. 1–42.
Grüneberg, W.J.; Ma, D.; Mwanga, R.O.M.; Carey, E.E.; Huamani, K.; Diaz, F.; Eyzaguirre, R.; Guaf, E.; Jusuf, M.; Karuniawan, A.; Tjintokohadi, K.; Song, Y.S.; Anil, S.R.; Hossain, M.; Rahaman, E.; Attaluri, S.I.; Some, K.; Afuape, S.O.; Adofo, K.; Lukonge, E.; Karanja, L.; Ndirigwe, J.; Ssemakula, G.; Agili, S.; Randrianaivoarinovy, J.M.; Chiona, M.; Chipungu, F.; Laurie, S.M.; Ricardo, J.; Andrade, M.; Rausch Fernandes, F.; Mello, A.S.; Khan, M.A.; Labonte, D.R.; Yencho, G.C. 2015. Advances in sweetpotato breeding from 1992 to 2012. In: Low, J.; Nyongesa, M.; Quinn, S.; Parker, M. (eds). Potato and sweetpotato in Africa. Transforming the value chains for food and nutrition security. Oxfordshire (UK). CABI International. ISBN 978-1-78064-420-2. pp. 3-68.
Huang, J. C., and Sun, M. 2000. Genetic diversity and relationships of sweetpotato and its wild relatives in Ipomoea series batatas (Convolvulaceae) as revealed by inter-simple sequence repeat (ISSR) and restriction analysis of chloroplast DNA. Theor. Appl. Genet. 100:10501060.
Lebot, V. 2010. Sweetpotato. In J.E. Bradshaw (ed.) Root and tuber crops. Vol 7. Springer Science & Business Media.
Low, J., J. Lynam, B. Lemaga, C. Crissman, I. Barker, G. Thiele, S. Namanda, C. Wheatley, and M. Andrade. “Sweetpotato in Sub-Saharan Africa.” In The sweetpotato, pp. 359-390. Springer Netherlands, 2009.
Mwanga, R., G.C. Yencho, R.W. Gibson, and J.W. Moyer. 2013. Methodology for inoculating sweetpotato virus disease: discovery of tip dieback, recovery and reversion in different clones. Plant Disease 97, 306.
Roullier, C, A. Duputie, P. Wennekes, L. Benoit, Fernandez Bringas VM, et al. 2013. Disentangling the Origins of Cultivated Sweet Potato (Ipomoea batatas (L.) Lam.). PLoS ONE 8(5): e62707. doi:10.1371/journal.pone.0062707
Yada, Benard, Phinehas Tukamuhabwa, Bramwell Wanjala, Dong-Jin Kim, Robert A. Skilton, Agnes Alajo, and Robert OM Mwanga. “Characterization of Ugandan sweetpotato germplasm using fluorescent labeled simple sequence repeat markers.” HortScience 45, no. 2 (2010): 225-230.
Zhang, D., J. Cervantes, E. Huaman, E. Carey, and M. Ghislain. 2000. Assessing genetic diversity of sweet potato (Ipomoesa batatas (L.) Lam.) cultivars from tropical America using AFLP. Genet. Resour. Crop Evol. 47:659665.

