Chapter 10: Ploidy: Polyploidy, Aneuploidy, and Haploidy
Deborah Muenchrath; Arden Campbell; Laura Merrick; Thomas Lübberstedt; Shui-Zhang Fei; and Walter Suza
Introduction
Not all plant species are diploids. In fact, 75% of all angiosperms are polyploids, which are characterized by having more than two sets of chromosomes in their somatic cells. About 70% of undomesticated grasses and 25% of legumes are polyploids. Among domesticated crop species in general, 75% have been found to be polyploid, with roughly similar percentages found for both annual and perennial species. Major crops, such as wheat, alfalfa, potato, cotton, and sugarcane, are polyploids. There are also plants that do not possess complete sets of chromosomes. Aneuploids have abnormal numbers of chromosomes and vary by the addition or deletion of specific individual chromosomes that otherwise would be present in the normal crop genome. Ploidy reduction produces haploids, which have only a single set of homologous chromosomes instead of the pair found in their diploid counterparts. Haploid plants are very valuable in certain breeding applications.
The number of chromosome sets possessed by a crop influences its genetics and thus, the strategies applied for its improvement. Plant breeders can alter chromosome numbers to modify and exploit genetic variability.
- Identify types and characteristics of polyploidy, aneuploidy, and haploidy.
- Understand how plants that vary in ploidy level occur naturally or are created artificially.
- Become familiar with the genetic behavior of polyploids, Aneuploids, haploids, and implications of changes in chromosome number for reproduction and productivity.
- Learn about the strengths and weaknesses of variation in ploidy level in plant breeding and genetic applications.
- Explore tissue and cell culture methods and their applications.
Concepts for Polyploidy
Ploidy
Polyploidy can be rather complex. The terminology and symbols used in connection with polyploidy communicate much information—understanding these will help clarify and simplify an otherwise complex topic.
- Ploidy refers to the number of chromosome sets in a cell. Prefixes are used to specify the number of chromosome sets in a particular organism. The symbol x is used to indicate the number of chromosomes in a set. Monoploids have one set (1x) and diploids have two sets (2x) of chromosomes, and so forth.
| Ploidy Level | Number of Chromosome Sets |
|---|---|
| Monoploid | 1x |
| Diploid | 2x |
| Triploid | 3x |
| Tetraploid | 4x |
| Pentaploid | 5x |
| Hexaploid | 6x |
| Heptaploid | 7x |
| Octoploid | 8x |
| Nonaploid | 9x |
| Decaploid | 10x |
Polyploid
- Polyploid is a general term indicating multiple (more than two) sets of chromosomes.
- Genome is a set of chromosomes that is inherited together, assuming normal meiosis and mitosis. Each genome is composed of the basic chromosome number, x, and each kind of chromosome is represented only once in each set.
Several symbols are used as shorthand to convey additional information about the chromosome constitution of the species or individual.
- n — indicates the haploid or gametic chromosome number of the species. (Note: n does not tell anything about the number of chromosome sets).
- 2n — denotes the diploid or somatic number of the species.
- x — indicates the basic number of chromosomes in each set or genome.
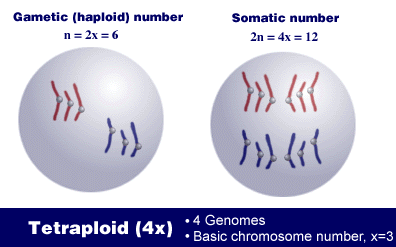
For example, a tetraploid, 4x, has 4 genomes or sets of chromosomes. The number of chromosomes in its somatic cells is 2n = 4x, and the number in its gametes is n = 2x. In this illustration, let three be the basic chromosome number, so x = 3. This means that each genome or chromosome set consists of 3 chromosomes.
Types of Polyploidy
Euploidy
There are two general types of ploidy, which include plants that have either one or more complete sets of chromosomes present in their genome (euploids) or those that have partial sets due to the absence of at least one of their individual chromosomes or presence of at least one extra one (aneuploids):
Euploidy refers to the number of chromosome sets in a cell. Prefixes are used to specify the number of chromosome sets in a particular organism. The symbol x is used to indicate the number of chromosomes in a set. Monoploids have one set (1x) and diploids have two sets (2x) of chromosomes, and so forth.
- Haploidy—individual with half (n=x) of the somatic cell chromosome number.
- Diploidy—individual with two sets of the basic, complete genome (2n=2x).
- Polyploidy—individual with more than two basic, complete sets of chromosomes in its somatic cells. Among polyploids, there are two main types:
- Autoploidy—individual has more than two complete chromosome sets from a single genome. Also known as autopolyploidy.
- Alloploidy—individual has two or more genomes contributed from different parental species in their ancestral lineage. Also known as allopolyploidy. Most naturally occurring polyploids are alloploids. Amphidiploids (also called amphiploids) are allotetraploids that contain two sets of genomes from each of two different parents. Amphidiploids are so-called because they behave like diploids during meiosis.
Autoploidy
Autoploids commonly occur as a result of duplication of the genome(s) of a single species, thus, the genomes possessed by an autoploid are identical. An autotetraploid (4x) has four sets of the same genome, e.g., four sets of the A genome (AAAA). The following are characteristics associated with autoploids:
- Greater ability to colonize new habitats than diploid ancestors—Due to gene buffering, autoploids often show a slower response to selection, but more adaptive potential.
- Dosage effect of gene expression—Additive effect of the alleles increases the number of phenotypes. There is a linear relationship between gene expression and number of gene copies.
- Larger cells and nuclei compared to their respective diploid counterparts—Because of the greater cell size, autoploids tend to have greater vegetative growth and produce larger structures. This feature has been utilized by breeders to increase yield of crops harvested for their vegetative structures.
- Reduced fertility—Abnormalities in meiosis may interfere with chromosome pairing, resulting in unbalanced chromosomal distribution between daughter cells, and thus, nonviable gametes. Gametes possessing an extra chromosome or missing a chromosome (i.e., aneuploids) are usually nonviable. This feature is utilized by breeders to develop seedless crops such as seedless watermelon.
Alloploidy
Alloploids arise when the genomes of two or more unrelated species are combined in a single individual.
Several characteristics are associated with alloploidy:
- Broadened genetic base. Alloploids (also known as amphiploids) behave like diploids and result in new species.
- Increased allele diversity and heterozygousity—Increase in possible allele combinations can provide expanded opportunities for breeding.
- Novel phenotypic variation—Genome interactions and changes in gene expression can occur in newly synthesized alloploids. Such gene changes can include transfer of sequences between genomes and gene conversion, loss, or silencing.
- Sterile unless genomes are doubled and chromosomes pair correctly. Chromosome doubling also occurs spontaneously in nature, mainly through the fusion of unreduced gametes.
Alloploidy has impacted the evolution of some major crops, such as wheat, cotton, tobacco, and various Brassicas (mustards). Information about the ploidy relationships between certain species has improved our understanding of the genetic origin of polyploid crops and facilitated their improvement.
In addition to identifying the genetic origin of a crop, genes, as well as whole genomes, can be transferred among related species to obtain novel genotypes and to combine the favorable qualities of different species as is demonstrated in the creation of Triticale.
Colchicine is a chemical that is commonly used to artificially double chromosomes. Colchicine is a toxic natural product that is found in the bulbs of autumn crocus (Colchicum autumnale). It is used as a medicine to treat the human disease gout, but it is also used to induce polyploidy in plants. Originally it was extracted from crocus bulbs, but is now also manufactured synthetically. When plant seeds or meristem tissues (apical tip, shoots, or suckers) are soaked in colchicine, it makes the cell walls permeable and prevents normal chromosome division.
Colchicine functions as a disruptor of mitosis by inhibiting microtubule formation, thus preventing normal chromosome segregation to occur. After seed treatment with the chemical, half the gametes resulting from meiosis contain no chromosomes, while the other half contains diploid instead of the normal haploid number. This disruption leads to embryos with double the usual number of chromosomes. Colchicine is used to make infertile hybrids or haploid plants fertile by restoring doubled chromosomes. The latter plants are known as doubled haploids. An example of the former is development of hexaploid and octaploid triticale.
Colchicine is also used in the study of karyotypes, which refer to the chromosome constitution of the cell (specifically, the number and morphological appearance of chromosomes in the nucleus). In order to observe chromosomes under a light microscope, cells are treated with the chemical near the middle of mitosis during metaphase—a point in the cell cycle when chromosomes are most dense and therefore most visible. Colchicine treatment arrests these chromosomes by preventing the formation of the spindle microtubules.
Wheat is an example of a crop whose origin has been extensively studied and is a classic example of a polyploid crop that evolved through the combination of genomes from related species. Wheat has three sets of chromosomes, designated as A, B, and D. Each of these sets contributes 7 chromosomes.
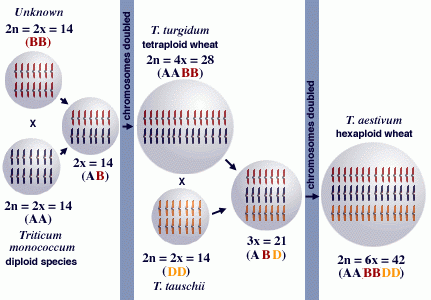
- wheat = AABBDD
- 1A, 2A, …, 7A;
- 1B, 2B, …, 7B;
- 1D, 2D, …, 7D.
The “Triangle of U” is a theory that the genomes of three ancestral species of Brassica combined through interspecific hybridization. Brassica species include both vegetable and oilseed crops, such as cabbage, broccoli, rapeseed, and various mustards.
The original theory was developed by Woo Jang-choon, a Korean botanist who published under the Japanese name Nagaharu U. He made artificial hybrids between closely related, but distinct diploid and polyploid species and examined how chromosomes paired in the progeny. Woo’s theory was later confirmed by examination of DNA from the group of related Brassica species.
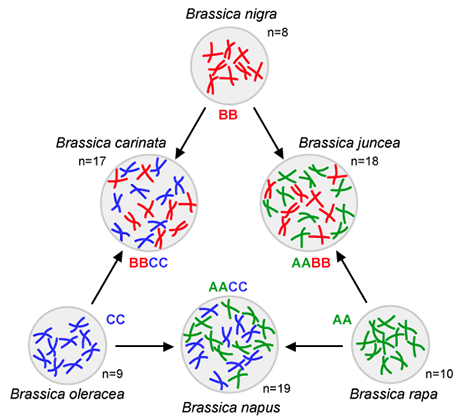
The different colors in the chromosomes depicted here represent the three ancestral genomes:
Genomes of three diploid species
AA: 2n = 2x = 20—Brassica rapa (syn. Brassica campestris) – turnip, Chinese cabbage
BB: 2n = 2x = 16—Brassica nigra – Black mustard
CC: 2n = 2x = 18—Brassica oleracea – cabbage, kale, broccoli, Brussel sprouts, cauliflower
AABB: 2n = 4x = 36—Brassica juncea—Indian mustard
AACC: 2n = 4x = 38—Brassica napus—Rapeseed, rutabaga
BBCC: 2n = 4x = 34—Brassica carinata—Ethiopian mustard
Autoploids and Alloploids
Many important crops are naturally occurring polyploids—either autoploids or alloploids. Letters are often used to indicate to the different source species that have been identified as contributors to crop genomes. However the same letter does not denote the same genome unless the species are closely related evolutionarily. For example, tall fescue (an alloploid) and perennial ryegrass (a diploid) both share the L genome in common. Pima and Upland cotton (both alloploids) share A and D genomes in common, but only share the A genome with two other cultivated cottons, Levant and Tree cotton (both diploids).
Other polyploid crops include sugarcane, yam, white potato, orchardgrass, highbush blueberry, persimmon, basil, nutmeg, and some types of asparagus and celery. Many horticultural species of flowers and ornamentals are polyploids. Some of these polyploids occur naturally and others were manipulated into polyploids through plant breeding efforts. Two examples of artificially created alloploids that are produced commercially include the cereal grain triticale (Tritcosecale) and the vegetable crop known as radicole (Raphanobrassica). Both were developed by intergeneric hybridization among closely related crop genera: wheat-rye and radish-cabbage (or radish plus other Brassicas).
Recent studies involving molecular techniques have revealed that many diploid crops with high chromosome numbers (2n>18-20) are actually paleopolyploids—plants whose genomes result from ancient genome duplications that occurred at least several million years ago. Known paleopolyploids of either alloploid or autoploid origins include such crop species as soybean, squash, maize, and lettuce.
Diploidization
In a process known as diploidization over time polyploid genomes may return to diploid status, although certain genes may be retained in multiple copies. Conversely, the term polyploidization refers to cycles of hybridization and chromosome doubling that result in either allopolyploids or autopolyploids.
| Crop Type | Diploid or Polyploid | Genome | Ploidy (x) |
|---|---|---|---|
| African rice | Diploid | A’A’ | 2x |
| Asian rice | Diploid | AA | 2x |
| Bread wheat | Alloploid | AABBDD | 6x |
| Durum wheat | Alloploid | AABB | 4x |
| Rye | Diploid | RR | 2x |
| Oats | Alloploid | AACCDD | 6x |
| Broccoli, Cabbage, Kale, Cauliflower, Brussel sprouts | Diploid | CC | 2x |
| Turnip, Chinese cabbage | Diploid | AA | 2x |
| Rapeseed, Rutabaga | Alloploid | AACC | 4x |
| Black Mustard | Diploid | BB | 2x |
| Brown Mustard | Alloploid | AABB | 4x |
| Ethopian Mustard | Alloploid | BBCC | 4x |
| Radish | Diploid | RR | 2x |
| Radicole | Alloploid | RRCC, RRAA | 4x |
| Sweet potato | Autoploid | BBBB, BBBBBB | 4x, 6x |
| Pima cotton | Alloploid | AADD | 4x |
| Tree cotton | Diploid | AADD | 4x |
| Upland cotton | Alloploid | AADD | 4x |
| Levant cotton | Diploid | AA | 2x |
| Safflower | Diploid | BB | 2x |
| Alfalfa | Autoploid | SSSS | 4x |
| Bermuda grass | Diploid, Autoploid | AA, AAAA | 2x, 4x |
| Tall fescue | Alloploid | PPLLXX | 6x |
| Perenial rye grass | Diploid | LL | 2x |
| Timothy | Alloploid | AAAABB | 6x |
| Banana | Diploid, Alloploid, Autoploid | AAA, AAA, ABB, AB, AAAB, AABB, ABBB | 2x-4x |
| Plum, European | Alloploid | CCSSSS | 6x |
| Coffee | Alloploid | AABB | 4x |
| Tobacco | Alloploid | SSTT | 4x |
| Crop Type | Hybrid or Parent | Genome |
|---|---|---|
| Triticale | Intergeneric hybrid | AABBRR, AABBDDRR |
| Bread wheat | Parental species | AABBDD |
| Durum wheat | Parental species | AABB |
| Rye | Parental species | RR |
| Radicole | Intergeneric hybrid | RRCC, RRAA |
| Broccoli, Cabbage, Kale, Cauliflower, Brussel sprouts | Parental species | CC |
| Turnip, Chinese cabbage | Parental species | AA |
| Turnip, Chinese cabbage | Parental species | RR |
Aneuploidy
Aneuploidy — organism has a partial set of chromosomes due to addition or deletion of specific chromosome(s). or one or more chromosome sets. It is a state in which the number of chromosomes is not an exact multiple of the haploid number.
There are numerous possible kinds of aneuploidy.
Aneuploids are described relative to the euploid condition. The general genomic formula for aneuploids is somatic chromosome number plus or minus the number of extra or missing chromosomes relative to the euploid somatic number: 2n ± one or more chromosomes. The following table describes the classification system used for aneuploids which includes not only the addition or absence of individual chromosomes (as described in the table below), but also includes the presence or absence of chromosomes characterized by partial rearrangements.
| Kind of Aneuploid | Genomic Formula | Interpretation Relative to Euploid Condition |
|---|---|---|
| Nullisomic | 2n – 2 | Euploid minus one pair of chromosomes |
| Monosomic | 2n – 1 | Euploid minus one chromosome |
| Trisomic | 2n + 1 | Euploid plus one chromosome |
| Tetrasomic | 2n + 2 | Euploid plus one pair of chromosomes |
How has aneuploidy been useful in plant breeding? Breeders and geneticists use aneuploids as a tool to
- identify the chromosomal location of specific genes, or
- substitute a particular chromosome into a genotype.
Chromosome transfer or substitution provides an efficient method to transfer specific characters without accompanying adverse traits. It also facilitates chromosome transfer from alien species, enabling the recipient population to acquire genes that are unavailable within its own gene pool. Aneuploids can be used to increase genetic diversity.
Induction of Polyploids, Aneuploids, and Haploids
Natural Induction of Polyploids
Polyploidy occurs spontaneously in nature. There are two mechanisms for natural induction of polyploidy.
- Unreduced gametes—When the chromosome number fails to be reduced during meiosis, unreduced gametes result and can produce polyploids if the unreduced gamete is successfully fused with reduced or unreduced gametes of the opposite sex.
| Female Gamete X | Male Gamete ⇒ | Resulting Zygote |
|---|---|---|
| n (reduced) | n (reduced) | 2n (normal) |
| 2n (unreduced) | n (reduced) | 2n + n (1 additional set of chromosomes from female) |
| n (reduced) | 2n (unreduced) | n + 2n (1 additional set of chromosomes from male) |
| 2n (unreduced) | 2n (unreduced) | 2n + 2n (2 additional sets of chromosomes, with 1 additional set from each parent) |
- Somatic chromosome doubling—Normal gamete fusion occurs but the chromosomes spontaneously double in the zygote or other somatic tissue. Such chromosome doubling is rare.
Ordinarily, conventional crosses between parents having different genomes (referred to as a “wide cross”) are unsuccessful. Their offspring usually exhibit:
- Reduced fertility as a result of problems in chromosome pairing during cell division.
- Reduced seed set.
- Reduced viability associated with genomic instability, especially in those with an odd number of chromosome sets (e.g., triploids, pentaploids).
Chromosome doubling generally restores or improves genetic stability, viability and fertility.
Artificial Induction of Polyploids
Polyploidy can also be induced artificially. Chemicals such as colchicine or environmental shocks can be used to disrupt normal chromosome division and induce polyploidy. Meristematic tissues are especially susceptible to such disruptions.
Induction of Aneuploids
Aneuploidy occurs during cell division when chromosomes do not segregate properly into daughter cells. When meiosis occurs, germ cells divide to create male and female gametes, typically with each sperm and egg ending up with the same number of chromosomes. However, when nondisjunction occurs, an extra copy may end up in one gamete and the other may not have any copies. In animals, aneuploidy is usually fatal. Plants can tolerate higher levels of aneuploidy, but aneuploidy female gametes are typically more viable than male gametes.
Triticale is a combination of wheat and rye. Efforts to mate these two crops began in 1888. Why was triticale developed? Triticale (Triticosecale) merges the desirable qualities of wheat and rye.
- Wheat possesses favorable flour and baking qualities and flavor.
- Rye contributes to improved vigor. Rye is winter hardy, adaptable to poorer soils and growing conditions, and resistant to some diseases.
Hexaploid triticale was created by combining the genomes of a tetraploid wheat (Triticum turgidum) with rye (Secale cereale). Rye is diploid. In this schematic, the wheat genomes are represented as A and B and the rye genome by R.
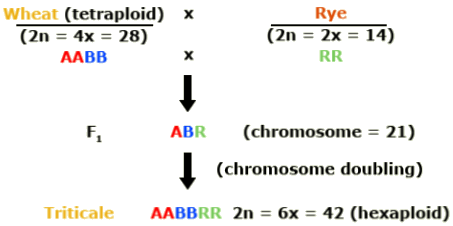
Octaploid triticale was formed through combination of hexaploid wheat (Triticum aestivum) and rye. The additional wheat genomes are represented by D in this illustration.
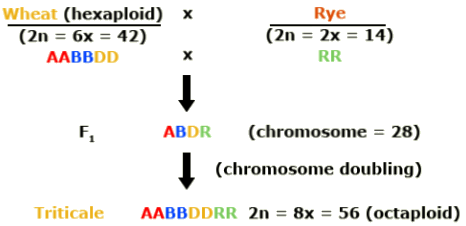
In the development of hexaploid and octaploid triticale, colchicine was used to double the chromosome number in the newly synthesized interspecific hybrids. Duplication of the genome resulted in restored fertility due to normal pairing of chromosomes in meiosis. With subsequent breeding, triticale was further improved. Today it resembles wheat, except that it has more vigorous growth and produces larger spikes and kernels.
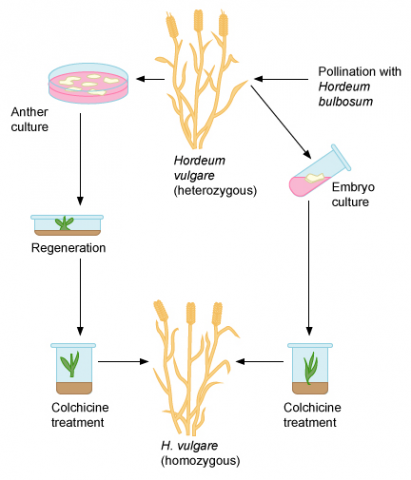
Induction of Haploids
Spontaneous haploid plants occur at low frequency in many species. In maize, for example, it occurs in the order of less than 1:1000. Haploid plants are of great interest in plant breeding, because genome doubling of haploids by chemical treatment (e.g., by use of colchicine) is possible, which leads to doubled haploid (DH) plants and lines, in which all loci are homozygous. Thus the development of inbred lines can be achieved in a much shorter time than by continued self-pollination starting from a heterozygous plant. To efficiently use DH lines, the spontaneous rates of haploid induction are not sufficient. Thus, procedures have been developed to induce haploidy.
Generally, haploids can be:
- maternal (based on the egg cell)
- paternal (based on microspores / pollen)
Moreover, haploids can be induced by using:
- Tissue culture techniques, such as anther or pollen culture.
- In vivo pollination techniques.
In maize, inducer genotypes are available for both paternal and maternal haploids. Current maternal inducer genotypes, when used as pollinator, result in 10% haploid offspring on the female parent. Respective inducer genotypes are available in other crops such as barley and potato. For some crops, other species are used as inducer. In case of wheat, for example, maize is used for induction of haploids.
Genetics of Polyploidy
Genetic Concepts
Polyploid genetics—especially autoploid genetics—is often more complicated than diploid genetics because genetic ratios are more complex. The inheritance of quantitative characters in polyploids is even more complicated. Before examining the complexity of polyploid genetics, let’s become familiar with terminology and associated principles.
- homoeologous chromosomes = corresponding chromosomes originating from different but similar genomes. Do not confuse homoeologous chromosomes with homologous chromosomes.
| Feature | Homoeologous | Homologous |
|---|---|---|
| Synapse of pair during the first division of meiosis? | Sometimes, but only at homologous or partially homologous regions | Yes |
| Have corresponding loci? | Sometimes | Yes |
| Derived from same genome? | No | Yes |
Valency
- Valency = the number of homologous or partially homologous chromosomes that associate or pair during meiosis. A prefix is added to indicate the number of chromosomes that associate. Here are some examples.
| Valency | Number of Meiotically Paired Chromosomes |
|---|---|
| Univalent | Single |
| Bivalent | 2 |
| Trivalent | 3 |
| Quadrivalent | 4 |
| Pentavalent | 5 |
| Hexavalent | 6 |
Polyploid genotypes can be defined by the
- number of dominant alleles at a locus, or
- number of different alleles at a locus.
Chromosomes
In each of the following examples, note the valency during meiosis and the chromosome constitution of the gametes. Think about what happens when these gametes fuse with normal or other abnormal gametes.
Example 1: Diploid
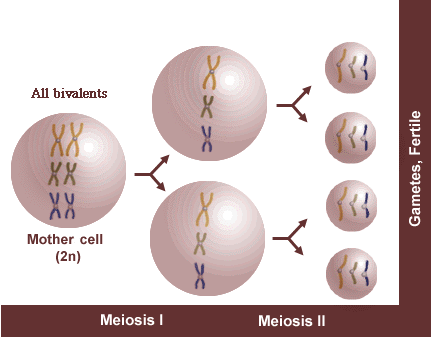
If meiosis proceeds normally, each resulting gamete will receive a full complement of chromosomes and be fertile. In this diploid, assume x = 3, n = 3, and 2n = 2x = 6.
Chromosomes form bivalents during meiosis I. One chromosome of each homologous pair migrates to a pole during anaphase I, resulting in a complete chromosome complement in each of two nuclei at the beginning of meiosis II. At anaphase II, sister chromatids separate resulting in a complete chromosome complement in each gamete—gametes are fertile.
Example 2: Autotetraploid
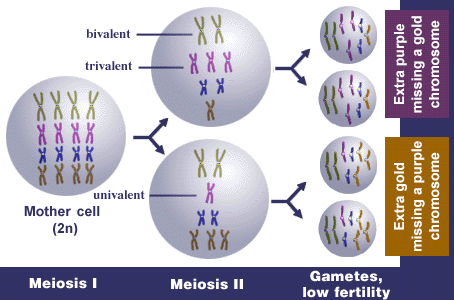
Normal synapsis in autotetraploids is as quadrivalents, e.g., homologues from all four genomes synapse. Normal first division separation is that bivalents go to each pole, and thus each gamete contains 2x. Random associations of bivalents can also occur. If homologous or partially homologous chromosomes fail to pair properly or fail to disjoin properly during meiosis, there is more likely to be unbalanced distribution of chromosomes to the daughter cells—some gametes will receive an extra chromosome and others will lack a chromosome, usually resulting in non-viable gametes. Thus, fertility is reduced. Reduced fertility is more common in autoploids than alloploids. Inheritance in autoploids is known as polysomic inheritance (or tetrasomic inheritance specifically in autotetraploids) and does not follow typical Mendelian patterns.
In this autotetraploid, let x = 4, n = 8, and 2n = 4x = 16.
Example 3: Allotetraploid.
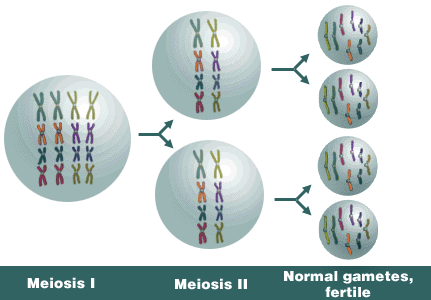
Alloploids generally have greater fertility than do autoploids. The formation of bivalents during meiosis contributes to greater fertility. Assume this allotetraploid has x = 4, n = 8, and 2n = 4x = 16.
Alloploid genetics is complex, too, but usually less so than in autoploids. The formation of bivalents in alloploids contributes to their less complex genetics. In addition, bivalency tends to result in disomic inheritance patterns, where two alleles segregate at a locus. Such inheritance of each individual duplicated loci follows typical Mendelian patterns. Thus, the genetic ratios of alloploids more closely resemble those commonly observed in diploids and it’s the reason why alloploids are often called amphiploids.
Alloploids that are derived from divergent progenitors exhibit disomic inheritance and are sometimes called genomic alloploids. Segmental alloploids are derived from partially divergent progenitors and typically exhibit mixed inheritance, consisting of both disomic and polysomic inheritance.
Select the most likely outcome when these gametes are fused with gametes having the chromosomal constitution indicated. In this question, Gamete A refers to the two upper types of gamete in the previous schematic, and Gamete B refers to the two lower ones.
Number of Dominant Alleles
The terms used to describe polyploid genotypes encode much information. Let’s define genotypes by the number of times a dominant (A) or recessive (a) allele is present at a particular locus. The most basic genotype is a nulliplex. A nulliplex is a polyploid in which all chromosomes of one homologous type carry the same recessive allele for the particular gene.
| Ploidy Level | Genotype |
|---|---|
| Diploid | aa |
| Triploid | aaa |
| Tetraploid | aaaa |
| … | |
| Octaploid | aaaaaaaa |
Other genotypes have at least one dominant allele. The number of dominant alleles is indicated by the term’s prefix. Let’s look at one locus of an autotetraploid as an example. There are five possible genotypes.
| Term | Genotype |
|---|---|
| Nulliplex | aaaa |
| Simplex | Aaaa |
| Duplex | AAaa |
| Triplex | AAAa |
| Quadruplex | AAAA |
If there is complete dominance, then all of the genotypes except the nulliplex will have the dominant phenotype. Only the nulliplex will exhibit the recessive phenotype. The presence of a dominant allele(s) can mask the presence of recessive alleles. The low frequency of recessive phenotypes necessitates the screening of larger populations in order to detect the presence of recessive alleles.
If there is partial dominance, the phenotypic ratios are very complex. Genetic linkages are extremely difficult to determine. It may even be difficult to identify autotetraploids as such because of intra- and interallelic interactions.
Number of Different Alleles
Let’s again use an autotetraploid and consider all the possible genotypes when there are four different alleles at a particular locus. We’ll designate these alleles as a1, a2, a3, and a4 (in this case, however, the use of small letters does not indicate recessive). An individual with all four different alleles present is called a “tetragenic.” Again, the terminology tells us much about the genetic composition.
| Term | Examples of Genotypes | Description |
|---|---|---|
| Nulliplex | a1a1a1a1, a2a2a2a2, a3a3a3a3, a4a4a4a4 | Only one allele present |
| Simplex | a1a1a1a2, a1a2a2a2, a3a1a1a1, a1a4a4a4, etc. | One allele present in three copies with another allele present in one copy |
| Duplex | a1a1a2a2, a1a1a3a3, a2a2a4a4, etc. | Two alleles, each present in two copies |
| Trigenic | a1a1a2a3, a2a2a3a4, a1a2a2a4, etc. | One allele present in two copies and two different alleles present in one each copy |
| Tetragenic | a1a2a3a4 | Four different alleles present |
In an autotetraploid, only four nulliplex genotypes are possible (a1a1a1a1, a2a2a2a2, a3a3a3a3, and a4a4a4a4), and only one tetragenic is possible (a1a2a3a4).
A tetraploid has multiple alleles (a1, a2, a3, and a4) at a particular locus. There is no dominance. A sample of the possible genotypes will be shown. You are required to match each of these genotypes to the term that best describes it. Some terms may have more than one genotype.
Autoploid Genetics
Now let’s take a closer look at why autoploid genetics is more complex than diploid genetics.
Multiple alleles, partial dominance, additive effects, and epistasis make the inheritance in autoploids more complex and linkage relationships “impossible” to decipher. Multi-allelism also affects the response to breeding procedure.
FURTHER THOUGHT
Many species of crop plants, as well as non-crop plants, are polyploids. Discuss the effects of polyploidy on plants and why this phenomenon is so prevalent.
Autotetraploid Genetics
Chromosomes assort independently in polyploids, just as they do in diploids. In addition, crossing-over can occur between homologous or partially homologous chromosomes. The higher the ploidy level, the more complex the genetics—the greater the number of chromosomes, the greater the number of possible combinations and genotypes that can be produced.
In an autoploid, the number of possible alleles is equal to the ploidy level. Thus, the higher the ploidy level, the greater the possible number of alleles for a given locus. Homologous chromosomes can each carry a different allele at a given locus. Recall that chromatids are the two strands comprising a duplicated chromosome that is still joined at the centromere. When the centromere divides—during anaphase of mitosis or anaphase II of meiosis—then chromatids separate and then are called chromosomes. Since chromosomes and chromatids assort randomly, the inheritance patterns are complicated.
Let’s examine the case of an autotetraploid. In this case, assume
- two alleles at the locus of interest: the “A” allele confers complete dominance and the “a” allele is recessive; and
- a duplex (AAaa) genotype.
Plants are self-pollinated and the segregation ratios of the progeny can be determined.
Follow the assortment of chromosomes through meiosis to determine the allelic composition of the resulting gametes.
Random Chromosome Assortment
Chromosome inheritance; two alleles, dominance, duplex (AAaa)
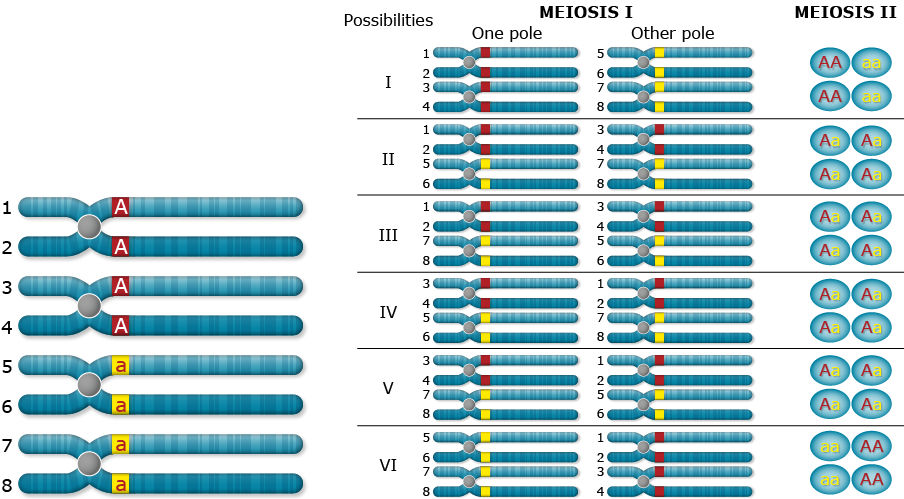
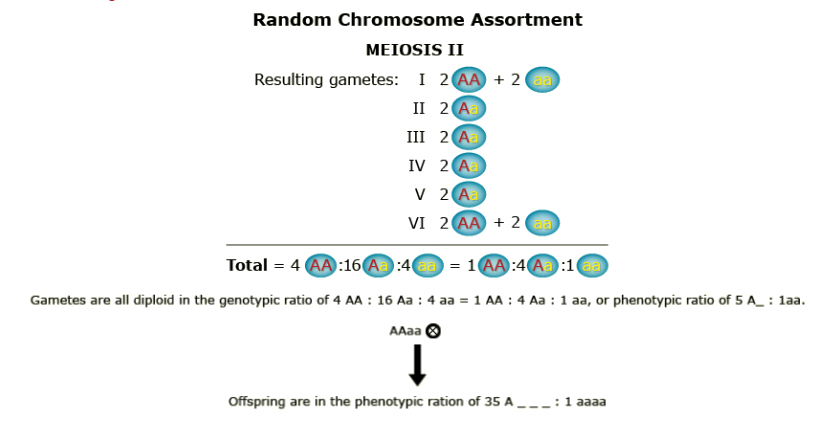
What happens when recombination occurs between the gene and the centromere? Let’s follow the random assortment of the chromatids through meiosis and find out the type of gametes produced.
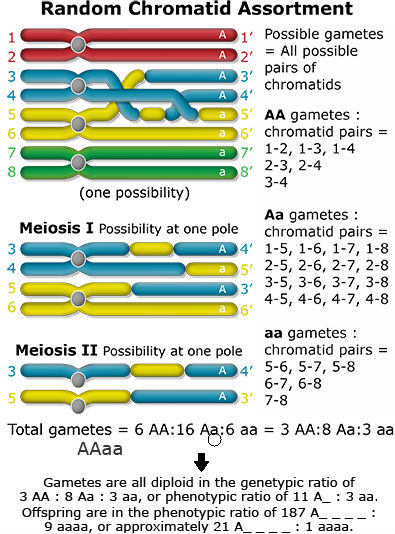
Conclusions
- Genetic segregation is more complex in autopolyploids compared to diploids. Homozygous recessive genotypes are more rare in autopolyploids than in diploids. Thus, if breeders intend to fix a particular allele, larger populations are required.
- There are different models describing genetic segregation in autopolyploids. Two extreme models have been mentioned above: chromosome and chromatid segregation. As shown for the duplex case AAaa, both models lead to different segregation rates, with homozygous recessives occurring at frequencies of 1/36 (chromosome segregation) and 1/22 (chromatid segregation). Which of these models is more accurate for a given gene, depends on the distance between this gene and the centromere. The larger the genetic distance, the more likely chromatid segregate independently.
Plant Cell and Tissue Culture
Plant cell and tissue culture methods enable plant breeders to obtain whole plants from somatic or haploid cells or tissues without sexual reproduction. Such methods are beneficial for handling plant material with variation in ploidy that may be otherwise difficult to reproduce. These methods take advantage of the totipotency of cells to generate plants.
Under special conditions, cells and tissues extracted from a donor plant can be induced to produce undifferentiated, unorganized cells that are totipotent. Totipotent cells possess all of the genetic information and capabilities needed to produce a whole organism. Because undifferentiated cells have not yet embarked on a developmental track to become a specific tissue or organ, their fate remains undetermined. These totipotent cells can be propagated in vitro and coaxed into developing roots and shoots, ultimately forming a whole plant.
Tissue culturing has several attractive features:
- Large numbers of totipotent cells can be cultured in a small area.
- The culture medium is usually well-defined—each component in the medium is known. Additions or exclusions of specific substances can provide a means of selection for some phenotypes.
- Ploidy level can be controlled by selection of explants.
- Heritable variation can be induced to generate new genotypes.
- Tissure culture is often required for genetic transformation.
General Tissue Culture Procedures
Cell and tissue culture procedures are similar. The particulars of this general procedure vary with the cell or tissue source, its age and health, species, genotype, culture type, and culturing objectives.
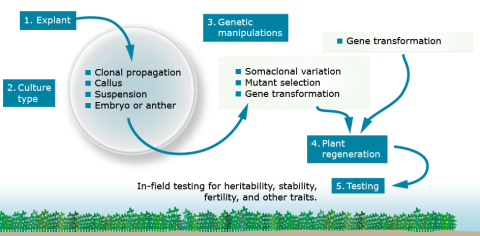
Types of Tissue Culture
Cell and tissue culture can be used to maintain genotypes or to obtain novel genotypes. Different tissues or types of culture are used to achieve a variety of objectives.
- Clonal propagation or Micropropagation—The general purpose of clonal propagation is to reproduce genetically identical plants without going through a gametophytic or sexual stage. A clone is a population of plants derived from a single plant, or a population of cells descendant from a single cell. This is very important in the production of high-value horticultural crops.
- Callus culture—Callus culture is similar to clonal propagation, except that the cultured tissue forms disorganized, undifferentiated masses of cells. Spontaneous variants and mutants can occur during callus culturing. Callus culture is often required for successful genetic transformation.
- Suspension culture—Similar to callus culture, except that cells or cell aggregates are in a liquid medium, rather than plated on an agar-solidified medium. These cultures have greater chromosome instability and reduced regeneration potential due to their rapid growth rates. The liquid medium provides relatively uniform exposure of cells to any selecting agents.
- Anther Culture—Anthers containing immature pollen are cultured to develop haploid plants or, with chromosome doubling, to obtain homozygous diploids. Anther culturing involves placing anthers containing immature pollen on medium. Generally, anthers are obtained from F1 or F2 plants to maximize the genetic diversity among the haploids generated through culturing. The greater the genetic diversity, the greater the likelihood of obtaining a desirable genotype to use as breeding lines.
- Embryo culture—Embryo culture allows the rescue of embryos that would normally be unobtainable or aborted because of some type of incompatibility. From the cultured embryos, whole plants may be regenerated.
Embryo culture is commonly used to bypass genomic incompatibility. Unfertilized embryos can be used to generate haploid plants or they can be pollinated in vitro to bypass self-incompatibility.
Genomic Incompatibility
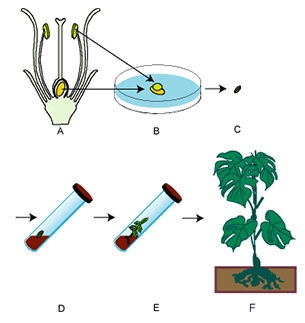
Often, embryos resulting from wide crosses (crosses between distantly related parents) are incompatible with the endosperm—the endosperm cannot adequately supply the needs of the growing embryo. As a result of this incompatibility, the embryo will normally abort before the seed is mature. Embryo culture provides a method to circumvent abortion and obtain such hybrids.
After fertilization, embryos are excised, with or without the ovule, and grown on a nutrient medium. The nutrient medium substitutes for the endosperm in nourishing the growing embryo. Via culturing, hybrid plants can be generated and grown to maturity. These plants will be sterile and must be treated with colchicine to get chromosome doubling. Flowers resulting from the doubled chromosome number will produce viable gametes much like allopolyploids.
Ovule Culture
Unfertilized ovules can be excised and cultured to obtain haploid plants. Alternatively, homozygous diploids can be generated by applying colchicine or other treatments to these cultured ovules to induce chromosomal doubling. This process is similar to anther culturing and will be discussed more fully in the next section. Ovule culturing has been used with barley, wheat, rice, and maize.
In vitro pollination and fertilization
In self-incompatibility systems where pollen tubes fail to grow, in vitro pollination offers an alternative to overcome such incompatibility. An unfertilized ovule is extracted from the pistillate flower and placed on medium. In vitro pollination and fertilization are accomplished by applying pollen directly to the ovule. Thus, the incompatibility factors localized on the stigma or in the style are avoided. The resulting embryo is nourished by the medium to obtain plantlets.
Uses of Polyploidy, Aneuploidy, and Haploidy
Because of the genetic complexities, breeding polyploids, especially autoploids, is challenging and progress is often slower than breeding diploids. Generally larger populations must be evaluated to obtain favorable types and to stabilize the genotype for the desired phenotype. Breeders also must be concerned about and test for the optimum ploidy level for the particular species and desired characters. Although polyploids present breeding challenges, they also offer opportunities for crop improvement.
ALLOPLOID USES
Alloploids occur more frequently in nature than other types of polyploids. Alloploids can combine the best characters of different species.
| Alloploid Uses | Example |
|---|---|
|
Wheat evolved through a combination of three related species. Cultivated Brassica species evolved through a series of interspecific hybridizations. |
|
Triticale resulted from a cross of wheat and rye. |
|
Cotton lint strength was improved through transfer of genes from undomesticated cotton into cultivated cotton.
(see Fehr, 1987, p. 82 “Introgression of genes”) |
Generally, alloploids are more vigorous and fertile than autoploids or aneuploids.
AUTOPLOID USES
Autoploids are not as common in nature as alloploids, but in some cases, they are developed as a result of artificially doubling the chromosome number of a diploid species. Depending on the crop, the reduced fertility that tends to occur as a result of induced polyploidy can be an advantage or a disadvantage. Reduced fertility found in autoploids is not a problem for species where the target organ is not seed—for example in forage crops. Fertility tends to be low and vegetative growth greater in autoploids than in diploids. This is an advantage in crops produced for vegetative parts; examples include several tuber and forage crops. Autoploidy is also used to generate sterile plants in which seeds are undesirable, such as in banana and watermelon.
Autoploids are also used to facilitate interspecific or wide crosses. Such crosses allow traits not otherwise available in the crop’s gene pool to be acquired, enhancing the genetic diversity of the crop.
ANEUPLOID USES
Aneuploids carry an extra chromosome(s) or are lacking one or more chromosomes — their genomes are unbalanced. Although this usually results in their reduced vigor, their unbalanced genomes provide benefits for genetic studies and increasing the genetic diversity of a crop. Diploids have very low viability in the aneuploid condition and, therefore, have limited utility. Aneuploidy is more useful in alloploids. Aneuploids have been used to:
- map genes to specific chromosomes
- identify linkage groups
- substitute or transfer chromosomes between related or unrelated species
With molecular techniques available today aneuploids are seldom used to map genes and identify linkage groups.
Chromosome transfer or substitution is a more efficient method than conventional crossing and subsequent segregation and selection because only the particular chromosome carrying the desired gene(s) is transferred. In conventional breeding approaches, both desired and undesired genes are transferred from the donor to the recipient population; subsequently, the desired and adverse traits must be sorted out, often requiring several generations of breeding and selection. Similar to the effects of wide crosses in alloploids, aneuploidy facilitates chromosome transfer from alien species, enabling the recipient population to acquire genes that are otherwise unavailable within its gene pool. Thus, aneuploids can be used to increase genetic diversity.
HAPLOID USES
Haploid plants are of interest in plant breeding and plant genetics because their genomes can be doubled and homozygous lines can be obtained much faster than by conventional techniques of inbreeding. Genetically homogeneous DH lines that can be quickly obtained make it practicable to establish respective experimental populations for gene mapping.
Discussion
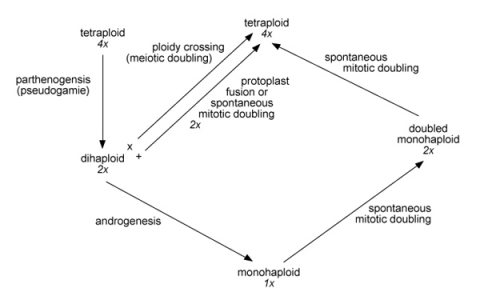
Wenzel proposed the analytic–synthetic breeding scheme in potato (shown below). Discuss the strengths and weaknesses of this scheme.
References
Becker, Heiko. 1993. Pflanzenzüchtung [Plant Breeding]. Ulmer & Co., Stuttgart, Germany.
Kuckuck, H., G. Kobabe, and G. Wenzel. 1991. Fundamentals of Plant Breeding. Springer Verlag, Berlin, Germany.
Ross, Hans. 1986. Potato breeding – problems and perspectives. In Fortschritte der Pflanzenzchtung [Advances in Plant Breeding]. Verlag Paul Parey, Berlin, Germany.
U, Nagaharu. 1935. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Japan. J. Bot. 7: 389-452.
An individual that has different alleles at a given locus, e.g., Aa or AAaa.
(1) The hereditary material of a cell comprising an entire chromosomal set found in each nucleus or organelle of a given species. See also organelle genome.
(2) The genome consists of a complete set of chromosomes (genes plus non-coding sequences) inherited as a unit from one parent.
One of the building blocks of DNA or RNA, each of which contains a nitrogenous base attached to a five-carbon sugar that in turn is attached to one or more phosphate groups.
A polyploid arising through the multiplication of the complete haploid chromosome set of a species.
The donor plants in the case of DH are the F1 or F2 progeny resulting from a breeding cross from which homozygous lines are desired.
Polyploid with genomes derived from different species.
Partial correlation displays the numerical relationship between a single Xi and Y in multiple correlation. In contrast to the simple correlation, the partial correlation accounts for interaction between the Xi's and illustrates the true relationship between the Xi and Y.
The change in allele frequency that occurs by chance (particularly in small populations) as a result of random sampling of the gametes that form the next generation.
Polyploid in which all chromosomes of one homologous type carry the same recessive allele for the particular gene.
A statistical artefact that is due to the deviation of estimates from true values by random error. In a mapping experiment, the loci that are deemed significant are enriched for those in which the estimated effects benefit from random error that happens to fall in the right direction. Therefore, significant QTLs are disproportionately those in which the effect sizes are inflated by chance.
A genotype formed when haploid cells undergo chromosome doubling.

