Chapter 10: Biotechnological Tools for Broadening Genetic Variation
Thomas Lübberstedt and Walter Suza
Crop breeding and genetic research both rely on genetic variation, which is synonymous with DNA variation. Therefore, the first step in a breeding program (Table 1) is, to generate genetic variation, from which superior genotypes can be selected. A critically important challenge in plant breeding is, to identify the best parents for establishing breeding populations, that have the highest chance of success to result in a superior variety. This is called “usefulness” of parent combinations.
In the most extreme form, genetic variation is not available in existing genotypes and thus warranting the need to create novel genetic variation. Traditionally, mutation breeding was used for this purpose by inducing mutants with chemicals or radiation. However, the entire process of mutation breeding is labor intensive. Now we can accomplish the same objective through manipulation of targeted genes in transgenic crop plants. As a result, genetically modified (GM) or biotechnology crops have set out on an unparalleled worldwide advance , and foods derived from GM crops are continuing to be approved for human consumption. With biotechnological tools becoming available, molecular cloning, transformation, and targeted introgression of transgenes into crop plants are used to generate genetic variation. The focus of this chapter will be on the application of biotechnological tools to produce genetic variation for crop breeding.
- Understand transformation, mutagenesis, and genome editing
- Understand position effect of transgenic events
- Understand the concept of Coexistence
- Familiarize with the concept of usefulness in parent selection
Application of Biotechnology Tools in Plant Breeding
New Variety Workflow
One of the important considerations in development of GM crops is the time lag between gene discovery and seed distribution to the farmers (Fig. 1). It takes about 15 years from identifying a relevant gene, to actually having it incorporated in a plant variety. This time lag is close to the timeframe needed to incorporate a new germplasm source into a commercial product. Thus, application of biotechnology in plant breeding must promise a significant improvement in yield or offer a useful, novel trait without generating yield drag.
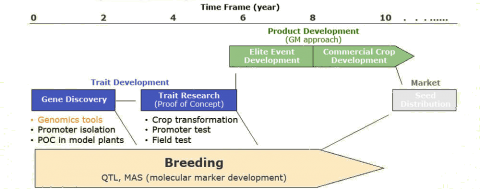
Relative Costs of Development
The process of creating a transgenic crop involves several steps, including gene discovery, promoter selection and testing, allele sequence modification for proper expression in plant cells, numerous transformation events, evaluation in crop plants at different stages, backcrossing into elite lines, production of experimental hybrids and varieties, and field testing. The last step is identification of elite events, which are transferred into the most recent germplasm. All these steps make the commercial development of transgenic varieties more costly as the development of varieties by conventional breeding (Table 2). For this reason, biotechnology is considered only an add-on to the actual breeding program, either conventional or by use of markers, which forms the basis for using those transgenes.
| Process |
EXOTIC | TRANSGENIC |
| Choice of Source/Discovery | 14,000 | 1,000,000 |
| Breeding/Modification | 38,000 | 100,000 |
| Efficacy Testing | 50,000 | |
| Transformation of Model Species | 50,000 | |
| Construct Comparisons | 50,000 | |
| Maize Transformation | 50,000 | |
| Backcrossing | 1,200 | |
| TOTAL COSTS | 52,000 | 1,301,200 |
Gene Stacking
Examples of biotechnology tools commonly used in plant breeding include gene stacking, nuclease-induced genome editing, artificial chromosomes, RNAi, transposon mutant collections, plant transformation, and TILLING. These tools are discussed in the following sections.
Gene stacking is a method of combining desired traits into a single line that has resulted in crops with several stacked-events (Table 2). The advantage of gene stacking is the benefit of obtaining a single seeds with several traits, for example, weed and pest resistance. This can be achieved through conventional breeding by mating two parents that each has a unique trait of interest. The main disadvantage of managing independently-segregating events is the large number of plants required to find at least one homozygous offspring. Consider an example of mating two parents each with a unique allele conferring a trait of interest, and using the doubled haploid (DH) technology to develop homozygous plants for the two desirable alleles. Even with DH technology, we see that about 100 plants must be screened to identify one with both alleles fixed (See Figure 16 in the module on Marker-Assisted Backcrossing). Thus, as alternative, transgenes can be cloned into a single construct (gene cassette), so that transgenes would co-segregate and inherited like a single gene. This would make MABC and handling of stacked genes much easier. Table 3 lists gene stacking technologies applied in different companies. These technologies will be the subject of discussion in the following sections.
Stacked Gene Examples
| Crop | Transgenic trait | Transgenic event(s) | Product name | Intended purpose | Developer |
|---|---|---|---|---|---|
| Maize | Cry1Ab, pat,
mutant maize EPSPS |
BT11, GA21 | Agrisure®
GT/CB/LL |
Lepidopteran pests
(European corn borer); Weeds |
Syngenta® |
| Maize | Cry1Fa, pat | TC1607 | Herculex®
CB |
Tolerance to European
corn borer; Weeds |
Dow®
AgroSciences and Pioneer® Hi-Bred |
| Maize | Cry1Ab, Cry3Bb1,
CP4 EPSPS |
MON810,
MON88017 |
Yeildgard®
VT Triple |
Tolerance to lepidopteran
and coleopteran insect pests; Weeds |
Monsato® |
| Canola | bar, barnase, barstar | MS8 (DBN230-0028),
RF3 (DBN212-005) |
Invigor®
SeedLink® |
Tolerance to weeds;
male sterility |
Bayer®
CropScience |
| Cotton | pat, Cry1Ac, CryFa | WideStrike® | Tolerance to weeds;
lepidopteran insect pests |
Dow®
AgroSciences |
However, transgene stacking may have some drawbacks. First, those genes of interest usually are not all available at once, but become available over a multitude of years. Thus, for the genes initially discovered, for which elite events have been identified already, the strategy would be to find elite events in the gene construct. By having two or more genes in a cassette, the likelihood of finding an elite event decreases because the two genes are essentially linked. The catch, however, is that if for some reason after some time one or more of the transgenes in a cassette are no longer of interest, the other transgene may also be rendered obsolete. In contrast, if the transgenes are independently segregating, then it is more flexible to combine or leave away transgenes that emerge over a longer period of time. Another issue is that stacking several transgenes may have a negative effect on the overall metabolism of the plant, and inadvertent reduction in yield.
Gene Stacking Technologies
| Technology | Developed by |
|---|---|
| Enzymes known as meganucleases are used to created stacked traits at genomic sites through homologous recombination | Cellectis |
| Application of protein engineering technology to develop meganucleases use din target-integration of transgenes in plant genomes | Precision Biosciences |
| Enzymes know as zin-finger nucleases (described in more details at later part of this module) are customized to fit specific needs | Sangamo Biosciences |
| Mini-chromosomes (described in more details in later parts of this module) | Chromatin, Inc. |
New Biotechnological Tools for Plant Transformation
Genome Editing
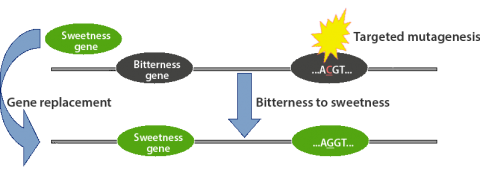
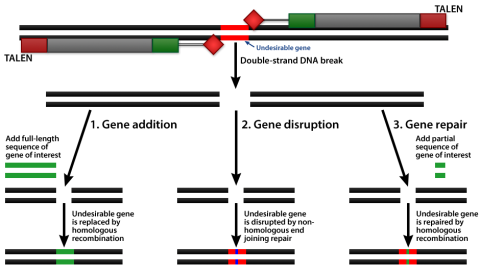
Genome editing with nucleases is a method used to cut desired locations in the genome to induce mutations to understand the function of genes or replace an endogenous gene with a novel allele or gene stacks (Fig. 2). In plants, nuclease-induced genome editing methods referred to as ZFNs and TALENs can be used for targeted introgression of stacked genes, allowing several physically linked traits to be inserted in a genomic region such that interference of the function of endogenous genes is avoided.
Zinc-Finger Nucleases (ZFNs)
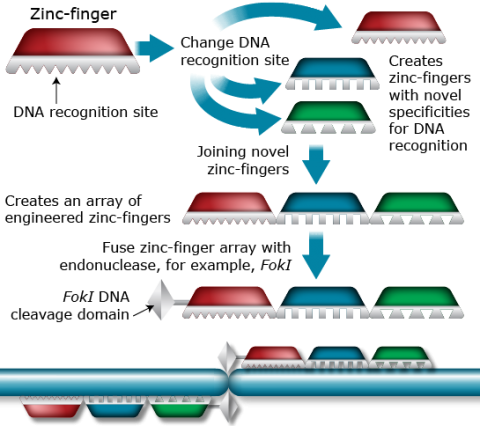
A zinc-finger is a DNA-binding domain of a protein that recognizes three base pairs of DNA. Engineered combinations of zinc fingers (Fig. 3) can be designed to bind longer stretches of DNA (in multiples of 3). Fusing a zinc-finger concatemer with a DNA-cleaving enzyme (nuclease), for example, the nuclease domain of the FokI restriction enzyme, results in “molecular scissors” that can modify specific DNA sequences recognized by a particular zinc-finger. However, a challenge with the ZFN technology is the low frequency of mutations which makes it difficult to identify the mutated alleles (Puchta and Hohn, 2010). Also, ZFNs have been known to produce off-target cleavage events.
Application of ZFN Technology
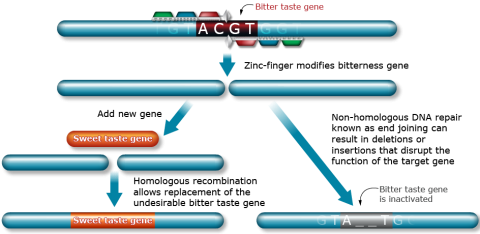
As mentioned earlier, ZFNs can be used to carry out site-directed mutagenesis in order to study gene function or replacing endogenous genes (Fig. 4)
TALENs
2. Transcription Activator-Like Effector Nucleases (TALENs)
TALENs are similar to ZFNs and comprise a non-specific FokI nuclease fused to a DNA binding domain (Fig. 5). The DNA-binding domain of a TALEN consists of highly conserved repeats of the transcription activator-like effectors (TALEs) from Xanthomonas spp. bacteria. TALEs are modular proteins that are composed of (i) an N-terminal translocation signal, (ii) a central DNA binding domain, and (iii) a C-terminal region containing nuclear localization and transcription activation signals. The TALE DNA binding domain consists of about 33-35 invariable repeat modules (Fig. 5A), with the exception of two hypervariable residues (referred to as repeat variable di-residues, RVDs) located at positions 12 and 13 (Fig. 5C). TALE repeats with different RVDs recognize different DNA base pairs (Fig. 5D). Consecutive RVDs in a TALE match directly the sequence of the DNA they bind, a characteristic referred to as the TALE code. Thus, the TALE code can be used to predict DNA target sequences. The simple relationship between RVDs sequence combinations and DNA binding specificity allows the engineering of novel DNA binding domains by selecting a combination of appropriate RVDs.
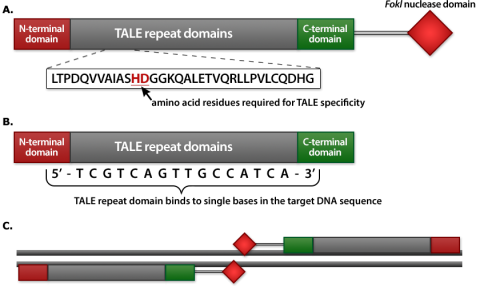
Fusion of TALEs
The biggest challenge with use of TALENs is engineering highly specific TALE domain to avoid off-target DNA cleavage. Such non-specific DNA editing may have deleterious results making it difficult to obtain a desirable mutation. Also, the RVD NN (Asparagine-Asparagine) has low specificity because it recognizes both guanine and adenine, whereas the guanine-specific RVD NK (asparagine-Lysine) does not function as well as NN. For these reasons, Seymour and Thrasher (2012) recommended the following TALE engineering strategies:
- Incorporation of at least 3-4 strong RVDs (e.g., HD or NN)
- Inclusion of position strong RVDs to avoid more than 6 stretches of weak RVDs, especially at the termini.
- Use of NH or NK for high guanine specificity.
- Use of NN for guanine if only a few other strong RVDs are present.
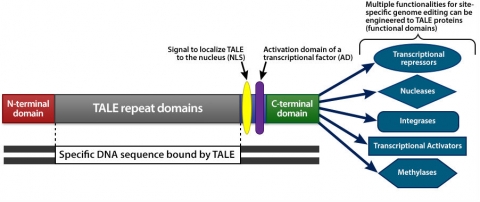
Application of TALEs
The DNA binding versatility of the TALE domain and the modular nature of these molecules allow their use for various purposes (Fig. 6 and 7). For example, they can be used to activate or repress gene expression, or edit the genome through nuclease activity to drive the replacement of endogenous DNA sequences with novel DNA sequences, and to mediate the integration of a transgene into native genome sequences.
Application of TALENs
CRISPR-Cas Gene Editing
Clustered regularly interspaced palindromic repeats (CRISPR)-CRISPR-associated (Cas) – CRISPR-Cas systems have gained prominence in animal and plant research. Combining the technology with genotype-independent plant transformation in crops such as maize can broaden the use of CRISPR-Cas and increase the speed and precision of crop improvement. Examples of traits that have been modified using CRISPR-Cas include higher resistance to powdery mildew in bread wheat, reduced breakdown of sucrose in potatoes during cold storage, and increased oleic acid content in soybean oil.
- This teaching resource explains the origin of the CRISPR-Cas system and its application in biotechnology.
- This YouTube video provides animation depicting the CRISPR-Cas9 method in genome editing.
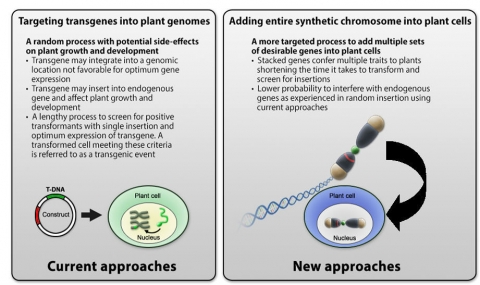
Artificial Chromosomes
Each Eukaryotic chromosome consists of a centromere and telomeres. The function of a centromere is to support spindle fibers when chromosomes segregate during meiosis and allow proper chromosome segregation. Telomeres consist of specific repeated DNA sequences and special proteins located at the tips of linear chromosomes. In addition to a centromere and telomeres, a plant’s artificial chromosomes must have an intact selectable marker and chromatin to allow replication (Fig. 9).
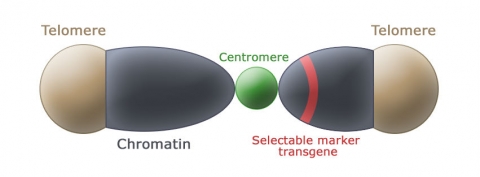
Advantages of Artificial Chromosomes
Although synthetic chromosomes in plants are still under research, they are likely to have more applications in the future. There are several advantages of using artificial chromosomes:
- They can be engineered to carry numerous transgenes (stacking) allowing many traits to be created at once (Fig. 10).
- Transgenes can be strategically placed in chromosomal regions that ensure they are expressed at a desirable level.
- Artificial chromosomes may be designed to contain specific recombination sites that would allow further additions of genes into a transgenic recipient of the artificial chromosome.
- Artificial chromosome could be introduced or removed by conventional genetic crosses.
RNAi
In some cases, decreased expression of an existing gene could be desired. For example, the content of a plant metabolite such as caffeine has to be reduced. RNA interference (RNAi) can be used to decrease the expression of genes through one of several different mechanisms including transcriptional silencing, translational silencing, or mRNA degradation.
RNAi can be accomplished in a more efficient way by expressing a portion of the target gene that has been engineered as an inverted repeat in transgenic crop plants. Following transcription of this engineered gene, the RNA molecules form a hairpin structure that is then cleaved into small fragments of double-stranded RNA, which interferes with the accumulation and function of the endogenous mRNA molecules of the target gene.
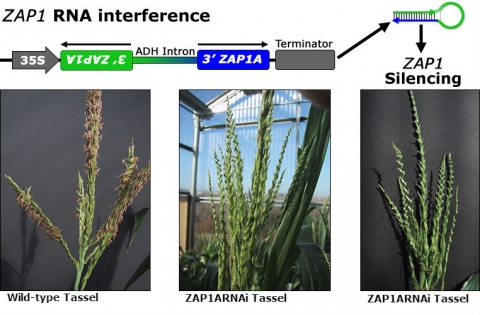
Male sterility is an important trait in hybrid seed production. To demonstrate the usefulness of RNAi in plant breeding let us consider induction of male sterility by reducing expression of key genes involved in floral development. The maize Zea Apetala1 (ZAP1) encodes a transcription factor that controls inflorescence architecture. The expression of ZAP1 is restricted to the sterile organs of the male floret (Mena et al. 1995). Consequently, RNAi silencing of ZAP1 results in male sterility (Fig. 11).
Transposon Mutant Collections
Transposable elements (TE) are DNA sequences found in all organisms that move from one location of the genome to another. If a TE inserts inside the coding or regulatory sequence of a gene, disruption of the gene can lead to a loss of gene function. Loss of gene function may result in obvious visible phenotypes. For this reason, TEs can provide useful reverse genetics strategy to determine function of genes discovered through current sequencing technologies. The creation of transposon mutant collections provides researchers additional tools to study gene function, and evolution of genomes.
Plant Transformation
Two commonly applied plant transformation procedures are Agrobacterium-mediated gene transfer and biolistics transformation (Fig. 12). Very few host cells receive the construct during the transformation process. Each random insertion of the construct into the genome of plant cells is referred as an event (transgenic event). Thus, an event is a unique DNA recombination event that takes place in a single plant cell, which is used to generate an entire transgenic plant (Fig. 12).

Transgenic Events
Not all transgenic events result on desirable expression of the transgene. Some events are poorly expressed because of position effects due to the nature of the site of chromosomal integration. A position effect is any transgene locus-specific effect generated by the insertion and/or expression of the gene (Fig. 13). In addition, transgenes can be inserted in multiple copies, or they can have undesirable pleiotropic effect, for example, by integration into an endogenous gene resulting and disrupting the function of such gene. In consequence, only a fraction of all events is considered as “elite events” for further evaluation in breeding materials.
Various Silencing Processes
Position effects may lead to transgene silencing through various processes, including DNA sequence modification by methylation, inhibition of mRNA processing, transport or translation, chromatin remodeling, and interactions between loci with homologous DNA sequences. The key question to ask before plant transformation, therefore, is how many independent events are needed? Fig. 14 shows program cascades for Maize (insect tolerance trait) and tomato (texture quality trait) at Monsanto and Syngenta, respectively. As seen in the figure, a large number of primary transformants needs to be screened to obtain stable events for improved texture and insect tolerance. Therefore, as a rule of thumb, >10 events are needed for testing constructs, and 50-100 events are needed for the final construct, to be certain to find at least one elite event.

Tilling
TILLING (Targeting Induced Local Lesions IN Genomes) is a biotechnological tool that employs chemical mutagenesis methods to create libraries of mutagenized seed that is later screened using high-throughput approaches for the discovery of useful mutations (Fig. 15). TILLING populations have been established to induce point mutations for subsequent forward and reverse genetic approaches, which in addition might contain novel and useful variants for breeding programs. Useful information about TILLING in rice, tomato, and Arabidopsis can be found at UC Davis’ Tilling website. With an increased understanding of sequence function relationships, valuable alleles might be identified in respective TILLING populations in a more targeted way by reverse genetic approaches.

Legal Considerations
Regulated Articles
In the US, all genetically modified plants are considered “regulated articles”. That means private and public institutions wishing to move or release a GM crop must obtain authorization (a notification or permit) from USDA Animal and Plant Health Inspection Service – APHIS.
The permit/notification must contain specific details about the genetic constitution, lineage, as well as testing and safety measures developed to ensure the GM crop is confined to the test site or is not maintained beyond the testing period. After years of field tests and evidence of low environmental risk, a certificate may be granted to grant a deregulated status to allow commercialization of the GM crop. After the GM crop has been deregulated it can be moved and sold to farmers. If the GM crop produces a compound that kills a pest (e.g., Bt maize and cotton) it is considered a pesticide and is subjected to regulation by the US Environmental Protection Agency (EPA). Information about EPA’s regulation of biotechnology for use in pest management can be found on the EPA’s Pesticides webpage.
Moreover, The US Food and Drug Administration (FDA) has regulatory powers over all food developed through the application of biotechnology. Thus, the complete procedure to register a variety developed by the use of biotechnology may cost between $6-15 million for a single event (Qaim, 2009). This has of course major implications for the use of transgenic approaches. Only those approaches that likely exceed the cost of regulatory approval in terms of return in investment may find their way to the market. This is one of the main reasons, that despite discovery of many gene candidates, actually only very few transgenes are used. Even though the area planted with transgenic crops has increased each year (James, 2008), the majority of the crops contain Bt and herbicide resistance traits. The high cost of developing transgenic crops suggests that only major private companies can afford to use those transgenes, and only few for major crops can be engineered with the transgenes.
GM Testing
Both public seed inspection bodies as well as private plant breeding/seed trading companies are carrying out systematic seed monitoring in order to detect possible admixtures of GM seeds as early as possible (see Markers and Sequencing). However, legal requirements may differ among countries, ranging from no requirements to mandatory use of event-specific quantitation. To ensure that countries abide by similar GM testing standards, the analytical methodology is harmonized at national and international levels (Table 4). For example, in addition to molecular data, other types of information are required in several countries that export or import GM crops (Table 5).
| Category of Information/Data Requirement | Argentina | Australia | Canada | Philippines | Japan | S. Africa | EU | USA |
|---|---|---|---|---|---|---|---|---|
| Copy number | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Number of insertion sites | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Organization of insert(s) | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Size of insert(s), complete &/ or partial | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Integrity genetic elements | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Absence of plasmid backbone | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Sequencing of T-DNA insert | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes |
| Sequencing of insert including 5′ & 3′ flanking | Information missing | Yes | Information missing | Information missing | Yes | No | Yes | Information missing |
| Bioinformatic analysis of novel ORFs and putative chimeric proteins | Information missing | Yes | Information missing | Information missing | Yes | No | Yes | Information missing |
| Sub-cellular location of insert(s) | Information missing | Information missing | Information missing | Information missing | Yes | Information missing | No | Information missing |
| Detection method | Information missing | Yes | Yes | Information missing | Yes | Yes | Yes | Information missing |
| Category of Information/Data Requirement | Argentina | Australia | Canada | Philippines | Japan | S. Africa | EU | USA |
|---|---|---|---|---|---|---|---|---|
| Host information | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Donor information | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Molecular information | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Molecular characterization | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Characterization of expressed protein | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Nutritional composition | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Potential toxicity of novel protein(s) | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Potential allergenicity of novel protein(s) | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
Coexistence
Coexistence as a choice refers to the ability of farmers to make a practical choice between conventional, organic and genetically modified (GM) crop production. As an issue, coexistence refers to the economic consequences of unintended presence of material from a GM crop in a non-GM crop and the principle that farmers should have a choice to freely produce agricultural crops they desire. As Fig. 16 shows, unintended entry of GM material into the non-GM pool can arise for a number of reasons. For example, seed impurities, cross pollination, volunteer crops, seed planting equipment, harvesting, transport, and storage and processing.

Application of Markers for Parent Selection
Successful Hybridization
Choice of parents of complementing parents (Table 1) is a critical task because it predetermines the result of the next phases in the breeding process and the allocation of resources in the breeding program. For this reason, markers are useful tools in assessing the genetic similarity among parents for prediction of the usefulness of a cross for line development.
Confirmation of Successful Hybridization
Molecular markers are useful in evaluating the success of hybridization of species that are not easy to visually verify whether seedlings were true hybrids, and those that require many years to flower.
For example, Clematis is a horticultural crop that takes 2-3 years to flower, and does not possess features that can be easily scored to select true hybrids. The application of RAPD and SNP markers has proven useful in verifying Clematis hybrids (Yuan et al., 2010).
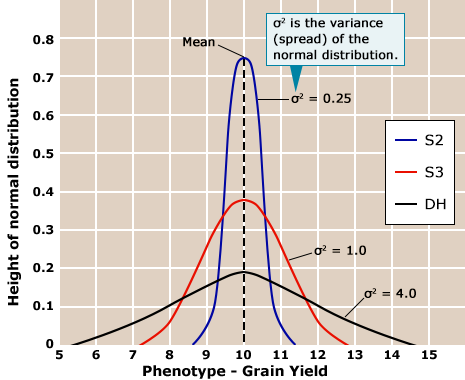
Usefulness Concept
Usefulness relates to a cross for line development and is defined as the sum of the population mean of all possible lines obtained from a cross in the absence of selection plus the predicted gain from selection. Therefore, usefulness depends on population mean and genotypic variance (Fig. 17). The following expression describes usefulness:
Applying the Usefulness Concept
The mean of a population can be reliably predicted based on the performance of the parental lines (Table 6). However, the remaining challenge is to predict the expected genotypic variance of a population. The genetic distance of the parental lines is a poor predictor (Table 6). Potentially genomic selection prediction (see Marker-Assisted Selection and Genomic Selection) will be a better alternative for this purpose in future. Currently, this is an area of active research.
| Predictor x | Heading date | Plant height | Lodging | Kernel weight | Grain yield | Sedimentation | Grain protein concentration |
|---|---|---|---|---|---|---|---|
| [latex]r(x, \hat{c}_{ij})[/latex] | |||||||
| Mean of parents [latex]\hat{m}_{ij}[/latex] | 0.90** | 0.90** | 0.76** | 0.79** | 0.74** | 0.71** | 0.37** |
| Mean of [latex]\textrm{F}_{2:4}[/latex] lines [latex]\hat{c}_{ij}[/latex] * | 0.90** | 0.93** | – | 0.67** | 0.52** | – | – |
| [latex]r(x, \hat{\sigma}^2_{g_{ij}})[/latex] ǂ | |||||||
| [latex]\textrm{P}\hat{\textrm{D}}_{ij}[/latex] ǂ | 0.22 | 0.32 | 0.35 | -0.17 | 0.18 | 0.02 | -0.11 |
| [latex]\textrm{P}\hat{\textrm{E}}_{ij}[/latex] ǂ | 0.12 | -0.13 | 0.22 | -0.25 | 0.21 | -0.26 | -0.11 |
| [latex]\textrm{var}(\textrm{F}_{2:4}\ \textrm{lines})[/latex] †ǂ | 0.59** | 0.59** | – | 0.52* | 0.08 | – | – |
| * Indicates significance at P = 0.05.
**Indicates significance at P = 0.01. † Phenotypic variance of line in Cross i x j. |
ǂ After logarithmic transformation was applied
[latex]\textrm{P}\hat{\textrm{D}}_{ij}[/latex] estimated phenotypic distance between Parents i and j for a given trait [latex]\textrm{P}\hat{\textrm{E}}_{ij}[/latex] estimated phenotypic Euclidean distance between Parents i and j. |
||||||
References
Bohn, M, H. F. Utz, and A. E. Melchinger. 1999. Genetic similarities among winter wheat cultivars determined on the basis of RFLPs, AFLPs, and SSRs and their use for predicting progeny variance. Crop Sci. 39: 228-237.
Brookes, G. Coexistence of GM and non GM crops: current experience and key principles.
Carlson, S. R., G. W. Rudgers, H. Zieler, et al. 2007. Meiotic transmission of an in vitro-assembled autonomous maize minichromosome. PLoS Genet 3: e179.
Carpenter, J. E. 2010. Peer-reviewed surveys indicate positive impact of commercialized GM crops. Nature Biotechnol. 28: 319-321.
Carroll, D. 2011. Genome engineering with zinc-finger nucleases. Genetics. 188: 773-782.
Cooper, J., B. J. Till, R. G. Laport, et al. 2008. TILLING to detect induced mutations in soybean. BMC Plant Biol. 8:9.
Curtin, S. J., F. Zhang, J. D. Sander, et al. 2011. Targeted mutagenesis of duplicated genes in soybean with zinc-finger nucleases. Plant Physiol. 156: 466-473.
Du, J., D. Grant, S. Tian, et al. 2010. SoyTEdb: a comprehensive database of transposable elements in the soybean genome. BMC Genomics 11: 113.
Ellen, L., G. van Enckevort, G. Droc, et al. 2005. EU-OSTID: A Collection of transposon insertional mutants for functional genomics in rice. Plant Mol. Biol. 59: 99-110.
Gaeta, R. T., R. E. Masonbrink, L. Krishnaswamy, et al. Synthetic chromosomes platforms in plants. Annu. Rev. Plant Biol. 63: 307-330.
Goodman, M. M. New sources of germplasm: Lines, transgenes, and breeders. In J. M. Martinez R., F. Rincon S, and G. Martinez G. (eds.). 2002. Memorial Congresso Nacional de Fitogenetica, Univ. Autonimo Agr. Antonio Narro, Saltillo, Coah., Mexico.
Holst-Jensen, A., M. De Loose, and G. Van den Eede. 2006. Coherence between legal requirements and approaches for detection of genetically modified organisms (GMOs) and their derived products. J. Agric. Food Chem. 54: 2799-2809.
James, C. 2008. Status of genetically modified crops: What is being grown, and where. Brief 39, Global Status of Commercialized Biotech/GM Crops: 2008, ISAAA.
Joung, J.K., and J.D. Sander. 2013. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 14(1):49-55. doi: 10.1038/nrm3486
Lusser, M., C. Parisi, D. Plan, and E. Rodríguez-Cerezo. 2012. Deployment of new biotechnologies in plant breeding. Nature Biotech. 30: 231-239.
Mahfouz, M.M., and L. Li. 2011. TALE nucleases and next generation GM crops. GM Crops: 2 (2), 99-103.
Mena, M., M. A. Mandel, D. R. Learner, et al. 1995. A characterization of the MADS-box gene family in maize. Plant J. 8: 845-854.
Osakabe, K., Y. Osakabe, and S. Toki. 2010. Site-directed mutagenesis in Arabidopsis using custom-designed zinc finger nucleases. Proc. Nat.l Acad. Sci. USA 107: 12034-12039.
Puchta, H., and B. Hohn. 2010. Breaking news: Plants mutate right on target. Proc Natl Acad Sci USA 107: 11657-116-58.
Qaim, M. 2009. The economics of genetically modified crops. Annu. Rev. Res. Econ. 1: 665-694.
Que, Q., M-D. M. Chilton, C. M. de Fontes, et al. 2010. Trait stacking in transgenic crops: Challenges and opportunities. GM Crops 1: 220-229.
Rommens, C. M., J. M. Humara, J. Ye, et al. 2004. Crop improvement through modification of the plant’s own genome. Plant Physiol. 135: 421-431.
Schouten, H.J., and E. Jacobsen. 2008. Cisgenesis and intragenesis, sisters in innovative plant breeding. Trends Plant Sci. 13: 260-261.
Stein, A. J., and E. Rodríguez-Cerezo. 2010. International trade and the global pipeline of new GM crops. Nature Biotechnol. 28: 23-25.
Streubel, J., C. Blücher, A. Landgraf, and J. Boch. 2012. TAL effector RVD specificities and efficiencies. Nature Biotechnol. 30: 593-595.
Till, B. J., S. H. Reynolds, C. Weil, et al. 2004. Discovery of induced point mutations in maize by TILLING. BMC Plant Biol. 4:12. doi:10.1186/1471-2229-4-12
Till, B. J., J. Cooper, T. H. Tai, et al. 2007. Discovery of chemically induced mutations in rice by TILLING. BMC Plant Biol. 7:19. doi:10.1186/1471-2229-7-19
Tolstrup, Karl; Andersen, Sven Bode, et al. Nov. 2003. Report from the Danish Working Group on the Co-existence of Genetically Modified Crops with Conventional and Organic Crops. Danish Institute of Agricultural Sciences report Plant Production no. 94.
Utz, H. F., M. Bohn, and A. E. Melchinger. 2001. Predicting progeny means and variances of winter wheat crosses from phenotypic values of their parents. Crop Sci. 41: 1470-1478.
Uauy, C., F. Paraiso, P. Colasuonno, et al. 2009. A modified TILLING approach to detect induced mutations in tetraploid and hexaploid wheat. BMC Plant Biol. 9:115. doi:10.1186/1471-2229-9-115
Yuan, T., L. Y. Wang, and M. S. Roh. 2010. Confirmation of Clematis hybrids using molecular markers. Scienti a Horticulturae 125: 136-145.
Zhong, S, and J-L. Jannink. 2007. Using quantitative trait loci results to discriminate among crosses on the basis of their progeny mean and variance. Genetics 177: 567-576.

